Comprehensive analysis of structural variants in breast cancer genomes using single-molecule sequencing
- PMID: 32887686
- PMCID: PMC7545150
- DOI: 10.1101/gr.260497.119
Comprehensive analysis of structural variants in breast cancer genomes using single-molecule sequencing
Abstract
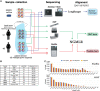
Improved identification of structural variants (SVs) in cancer can lead to more targeted and effective treatment options as well as advance our basic understanding of the disease and its progression. We performed whole-genome sequencing of the SKBR3 breast cancer cell line and patient-derived tumor and normal organoids from two breast cancer patients using Illumina/10x Genomics, Pacific Biosciences (PacBio), and Oxford Nanopore Technologies (ONT) sequencing. We then inferred SVs and large-scale allele-specific copy number variants (CNVs) using an ensemble of methods. Our findings show that long-read sequencing allows for substantially more accurate and sensitive SV detection, with between 90% and 95% of variants supported by each long-read technology also supported by the other. We also report high accuracy for long reads even at relatively low coverage (25×-30×). Furthermore, we integrated SV and CNV data into a unifying karyotype-graph structure to present a more accurate representation of the mutated cancer genomes. We find hundreds of variants within known cancer-related genes detectable only through long-read sequencing. These findings highlight the need for long-read sequencing of cancer genomes for the precise analysis of their genetic instability.
© 2020 Aganezov et al.; Published by Cold Spring Harbor Laboratory Press.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
