Telomere-to-telomere assembled and centromere annotated genomes of the two main subspecies of the button mushroom Agaricus bisporus reveal especially polymorphic chromosome ends
- PMID: 32887908
- PMCID: PMC7473861
- DOI: 10.1038/s41598-020-71043-5
Telomere-to-telomere assembled and centromere annotated genomes of the two main subspecies of the button mushroom Agaricus bisporus reveal especially polymorphic chromosome ends
Erratum in
-
Author Correction: Telomere-to-telomere assembled and centromere annotated genomes of the two main subspecies of the button mushroom Agaricus bisporus reveal especially polymorphic chromosome ends.Sci Rep. 2021 Aug 12;11(1):16734. doi: 10.1038/s41598-021-96123-y. Sci Rep. 2021. PMID: 34385579 Free PMC article. No abstract available.
Abstract
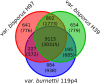
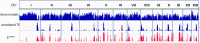
Agaricus bisporus, the most cultivated edible mushroom worldwide, is represented mainly by the subspecies var. bisporus and var. burnettii. var. bisporus has a secondarily homothallic life cycle with recombination restricted to chromosome ends, while var. burnettii is heterothallic with recombination seemingly equally distributed over the chromosomes. To better understand the relationship between genomic make-up and different lifestyles, we have de novo sequenced a burnettii homokaryon and synchronised gene annotations with updated versions of the published genomes of var. bisporus. The genomes were assembled into telomere-to-telomere chromosomes and a consistent set of gene predictions was generated. The genomes of both subspecies were largely co-linear, and especially the chromosome ends differed in gene model content between the two subspecies. A single large cluster of repeats was found on each chromosome at the same respective position in all strains, harbouring nearly 50% of all repeats and likely representing centromeres. Repeats were all heavily methylated. Finally, a mapping population of var. burnettii confirmed an even distribution of crossovers in meiosis, contrasting the recombination landscape of var. bisporus. The new findings using the exceptionally complete and well annotated genomes of this basidiomycete demonstrate the importance for unravelling genetic components underlying the different life cycles.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Mating-Type Locus Organization and Mating-Type Chromosome Differentiation in the Bipolar Edible Button Mushroom Agaricus bisporus.Genes (Basel). 2021 Jul 16;12(7):1079. doi: 10.3390/genes12071079. Genes (Basel). 2021. PMID: 34356095 Free PMC article.
-
An expanded genetic linkage map of an intervarietal Agaricus bisporus var. bisporusxA. bisporus var. burnettii hybrid based on AFLP, SSR and CAPS markers sheds light on the recombination behaviour of the species.Fungal Genet Biol. 2010 Mar;47(3):226-36. doi: 10.1016/j.fgb.2009.12.003. Epub 2009 Dec 21. Fungal Genet Biol. 2010. PMID: 20026415
-
Whole Genome Sequence of the Commercially Relevant Mushroom Strain Agaricus bisporus var. bisporus ARP23.G3 (Bethesda). 2019 Oct 7;9(10):3057-3066. doi: 10.1534/g3.119.400563. G3 (Bethesda). 2019. PMID: 31371382 Free PMC article.
-
Developments in breeding of Agaricus bisporus var. bisporus: progress made and technical and legal hurdles to take.Appl Microbiol Biotechnol. 2017 Mar;101(5):1819-1829. doi: 10.1007/s00253-017-8102-2. Epub 2017 Jan 28. Appl Microbiol Biotechnol. 2017. PMID: 28130632 Free PMC article. Review.
-
Critical Factors Involved in Primordia Building in Agaricus bisporus: A Review.Molecules. 2020 Jun 29;25(13):2984. doi: 10.3390/molecules25132984. Molecules. 2020. PMID: 32610638 Free PMC article. Review.
Cited by
-
Cycling in degradation of organic polymers and uptake of nutrients by a litter-degrading fungus.Environ Microbiol. 2021 Jan;23(1):224-238. doi: 10.1111/1462-2920.15297. Epub 2020 Nov 9. Environ Microbiol. 2021. PMID: 33140552 Free PMC article.
-
Lessons on fruiting body morphogenesis from genomes and transcriptomes of Agaricomycetes.Stud Mycol. 2023 Jul;104:1-85. doi: 10.3114/sim.2022.104.01. Epub 2023 Jan 31. Stud Mycol. 2023. PMID: 37351542 Free PMC article.
-
A genetic linkage map and improved genome assembly of the termite symbiont Termitomyces cryptogamus.BMC Genomics. 2023 Mar 16;24(1):123. doi: 10.1186/s12864-023-09210-x. BMC Genomics. 2023. PMID: 36927388 Free PMC article.
-
The First Whole Genome Sequencing of Agaricus bitorquis and Its Metabolite Profiling.J Fungi (Basel). 2023 Apr 18;9(4):485. doi: 10.3390/jof9040485. J Fungi (Basel). 2023. PMID: 37108939 Free PMC article.
-
Mating-Type Locus Organization and Mating-Type Chromosome Differentiation in the Bipolar Edible Button Mushroom Agaricus bisporus.Genes (Basel). 2021 Jul 16;12(7):1079. doi: 10.3390/genes12071079. Genes (Basel). 2021. PMID: 34356095 Free PMC article.
References
-
- Royse JM, May B. Use of isozyme variation to identify genotypic classes of Agaricus brunnescens. Mycologia. 1982;74:93–102. doi: 10.1080/00275514.1982.12021473. - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

