Telomere-to-telomere assembled and centromere annotated genomes of the two main subspecies of the button mushroom Agaricus bisporus reveal especially polymorphic chromosome ends
- PMID: 32887908
- PMCID: PMC7473861
- DOI: 10.1038/s41598-020-71043-5
Telomere-to-telomere assembled and centromere annotated genomes of the two main subspecies of the button mushroom Agaricus bisporus reveal especially polymorphic chromosome ends
Erratum in
-
Author Correction: Telomere-to-telomere assembled and centromere annotated genomes of the two main subspecies of the button mushroom Agaricus bisporus reveal especially polymorphic chromosome ends.Sci Rep. 2021 Aug 12;11(1):16734. doi: 10.1038/s41598-021-96123-y. Sci Rep. 2021. PMID: 34385579 Free PMC article. No abstract available.
Abstract
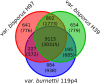
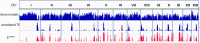
Agaricus bisporus, the most cultivated edible mushroom worldwide, is represented mainly by the subspecies var. bisporus and var. burnettii. var. bisporus has a secondarily homothallic life cycle with recombination restricted to chromosome ends, while var. burnettii is heterothallic with recombination seemingly equally distributed over the chromosomes. To better understand the relationship between genomic make-up and different lifestyles, we have de novo sequenced a burnettii homokaryon and synchronised gene annotations with updated versions of the published genomes of var. bisporus. The genomes were assembled into telomere-to-telomere chromosomes and a consistent set of gene predictions was generated. The genomes of both subspecies were largely co-linear, and especially the chromosome ends differed in gene model content between the two subspecies. A single large cluster of repeats was found on each chromosome at the same respective position in all strains, harbouring nearly 50% of all repeats and likely representing centromeres. Repeats were all heavily methylated. Finally, a mapping population of var. burnettii confirmed an even distribution of crossovers in meiosis, contrasting the recombination landscape of var. bisporus. The new findings using the exceptionally complete and well annotated genomes of this basidiomycete demonstrate the importance for unravelling genetic components underlying the different life cycles.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Royse JM, May B. Use of isozyme variation to identify genotypic classes of Agaricus brunnescens. Mycologia. 1982;74:93–102. doi: 10.1080/00275514.1982.12021473. - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

