A Cell-Autonomous Signature of Dysregulated Protein Phosphorylation Underlies Muscle Insulin Resistance in Type 2 Diabetes
- PMID: 32888406
- PMCID: PMC7875546
- DOI: 10.1016/j.cmet.2020.08.007
A Cell-Autonomous Signature of Dysregulated Protein Phosphorylation Underlies Muscle Insulin Resistance in Type 2 Diabetes
Abstract
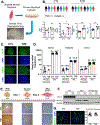
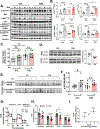
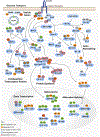
Skeletal muscle insulin resistance is the earliest defect in type 2 diabetes (T2D), preceding and predicting disease development. To what extent this reflects a primary defect or is secondary to tissue cross talk due to changes in hormones or circulating metabolites is unknown. To address this question, we have developed an in vitro disease-in-a-dish model using iPS cells from T2D patients differentiated into myoblasts (iMyos). We find that T2D iMyos in culture exhibit multiple defects mirroring human disease, including an altered insulin signaling, decreased insulin-stimulated glucose uptake, and reduced mitochondrial oxidation. More strikingly, global phosphoproteomic analysis reveals a multidimensional network of signaling defects in T2D iMyos going beyond the canonical insulin-signaling cascade, including proteins involved in regulation of Rho GTPases, mRNA splicing and/or processing, vesicular trafficking, gene transcription, and chromatin remodeling. These cell-autonomous defects and the dysregulated network of protein phosphorylation reveal a new dimension in the cellular mechanisms underlying the fundamental defects in T2D.
Keywords: chromatin remodeling; glucose transport; iPSC; insulin resistance; mRNA splicing; mitochondrial oxidation; phosphoproteomics; skeletal muscle; type 2 diabetes; vesicle trafficking.
Copyright © 2020. Published by Elsevier Inc.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures


References
-
- Al-Khalili L, Bouzakri K, Glund S, Lonnqvist F, Koistinen HA, and Krook A (2006). Signaling specificity of interleukin-6 action on glucose and lipid metabolism in skeletal muscle. Mol Endocrinol 20, 3364–3375. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

