Osteopontin Expression Identifies a Subset of Recruited Macrophages Distinct from Kupffer Cells in the Fatty Liver
- PMID: 32888418
- PMCID: PMC7501731
- DOI: 10.1016/j.immuni.2020.08.004
Osteopontin Expression Identifies a Subset of Recruited Macrophages Distinct from Kupffer Cells in the Fatty Liver
Abstract
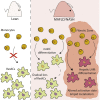
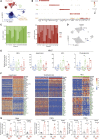
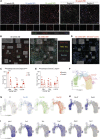
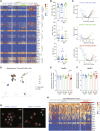
Metabolic-associated fatty liver disease (MAFLD) represents a spectrum of disease states ranging from simple steatosis to non-alcoholic steatohepatitis (NASH). Hepatic macrophages, specifically Kupffer cells (KCs), are suggested to play important roles in the pathogenesis of MAFLD through their activation, although the exact roles played by these cells remain unclear. Here, we demonstrated that KCs were reduced in MAFLD being replaced by macrophages originating from the bone marrow. Recruited macrophages existed in two subsets with distinct activation states, either closely resembling homeostatic KCs or lipid-associated macrophages (LAMs) from obese adipose tissue. Hepatic LAMs expressed Osteopontin, a biomarker for patients with NASH, linked with the development of fibrosis. Fitting with this, LAMs were found in regions of the liver with reduced numbers of KCs, characterized by increased Desmin expression. Together, our data highlight considerable heterogeneity within the macrophage pool and suggest a need for more specific macrophage targeting strategies in MAFLD.
Keywords: Kupffer cells; MAFLD; NAFLD; NASH; Western Diet; lipid; liver; macrophages; subsets.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures




Comment in
-
Two Roads Diverge in the Sick Liver, Monocytes Travel Both.Immunity. 2020 Sep 15;53(3):479-481. doi: 10.1016/j.immuni.2020.08.006. Immunity. 2020. PMID: 32937148
-
Macrophage Heterogeneity in NASH: More Than Just Nomenclature.Hepatology. 2021 Jul;74(1):515-518. doi: 10.1002/hep.31790. Epub 2021 May 22. Hepatology. 2021. PMID: 33666272 Free PMC article. No abstract available.
References
-
- Ballardini G., Fallani M., Biagini G., Bianchi F.B., Pisi E. Desmin and actin in the identification of Ito cells and in monitoring their evolution to myofibroblasts in experimental liver fibrosis. Virchows Arch. B Cell Pathol. Incl. Mol. Pathol. 1988;56:45–49. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

