Mortality Risk Profiling of Staphylococcus aureus Bacteremia by Multi-omic Serum Analysis Reveals Early Predictive and Pathogenic Signatures
- PMID: 32888495
- PMCID: PMC7494005
- DOI: 10.1016/j.cell.2020.07.040
Mortality Risk Profiling of Staphylococcus aureus Bacteremia by Multi-omic Serum Analysis Reveals Early Predictive and Pathogenic Signatures
Abstract
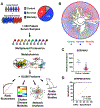
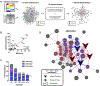
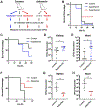
Staphylococcus aureus bacteremia (SaB) causes significant disease in humans, carrying mortality rates of ∼25%. The ability to rapidly predict SaB patient responses and guide personalized treatment regimens could reduce mortality. Here, we present a resource of SaB prognostic biomarkers. Integrating proteomic and metabolomic techniques enabled the identification of >10,000 features from >200 serum samples collected upon clinical presentation. We interrogated the complexity of serum using multiple computational strategies, which provided a comprehensive view of the early host response to infection. Our biomarkers exceed the predictive capabilities of those previously reported, particularly when used in combination. Last, we validated the biological contribution of mortality-associated pathways using a murine model of SaB. Our findings represent a starting point for the development of a prognostic test for identifying high-risk patients at a time early enough to trigger intensive monitoring and interventions.
Keywords: Staphylococcus aureus; bacteremia; biomarkers; host-pathogen interaction; infectious disease; metabolomics; post-translational modifications; proteomics.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests G.S. has received speaking honoraria from Allergan and Melinta Pharmaceuticals and consulting fees from Allergan and Paratek Pharmaceuticals
Figures

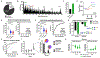


References
-
- Baxter RC (2014). IGF binding proteins in cancer: mechanistic and clinical insights. Nat Rev Cancer 14, 329–341. - PubMed
-
- Bello G, Pennisi MA, Montini L, Silva S, Maviglia R, Cavallaro F, Bianchi A, De Marinis L, and Antonelli M (2009). Nonthyroidal illness syndrome and prolonged mechanical ventilation in patients admitted to the ICU. Chest 135, 1448–1454. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI148417/AI/NIAID NIH HHS/United States
- F30 CA243480/CA/NCI NIH HHS/United States
- U01 AI124316/AI/NIAID NIH HHS/United States
- T32 AR064194/AR/NIAMS NIH HHS/United States
- R21 AI144060/AI/NIAID NIH HHS/United States
- R01 HL125352/HL/NHLBI NIH HHS/United States
- T32 DK007202/DK/NIDDK NIH HHS/United States
- T32 GM007752/GM/NIGMS NIH HHS/United States
- R01 AI141401/AI/NIAID NIH HHS/United States
- U54 HD090259/HD/NICHD NIH HHS/United States
- R01 AI132627/AI/NIAID NIH HHS/United States
- R21 AI149090/AI/NIAID NIH HHS/United States
- P50 HD090259/HD/NICHD NIH HHS/United States
- R01 AI144694/AI/NIAID NIH HHS/United States

