Assessing oligonucleotide designs from early lab developed PCR diagnostic tests for SARS-CoV-2 using the PCR_strainer pipeline
- PMID: 32889496
- PMCID: PMC7441044
- DOI: 10.1016/j.jcv.2020.104581
Assessing oligonucleotide designs from early lab developed PCR diagnostic tests for SARS-CoV-2 using the PCR_strainer pipeline
Abstract
Introduction: During the first month of the SARS-CoV-2 outbreak, rapid development of PCR-based diagnostic tests became a global priority so that timely diagnosis, isolation, and contact tracing could minimize the advancing pandemic surge. Designing these tests for broad, long-term detection was complicated by limited information about the novel virus' genome sequence and how it might mutate during global spread and adaptation to humans.
Methods: We assessed eight widely adopted lab developed PCR tests for SARS-CoV-2 against 15,001 SARS-CoV-2 genome sequences. Using a custom bioinformatic pipeline called PCR_strainer, we identified all mismatches and sequence variants in genome locations targeted by 15 sets of primer/probe oligonucleotides from these assays.
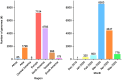
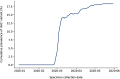
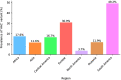
Results: For 12 out of 15 primer/probe sets, over 98 % of SARS-CoV-2 genomes had no mismatches. Two primer/probe sets contained a single mismatch in the reverse primer that was present in over 99 % of genomes. One primer/probe set targeted a location with extensive polymorphisms with 23 sequence observed variants at the forward primer location. One of these variants, which contains three nucleotide mismatches, arose in February as part of the emergence of a viral clade and was present in 18.8 % of the genomes we analyzed.
Discussion: Most early PCR diagnostic tests for SARS-CoV-2 remain inclusive of circulating viral diversity, but three assays with extensive mismatches highlight assay design challenges for novel pathogens and provide valuable lessons for PCR assay design during future outbreaks. Our bioinformatics pipeline is also presented as a useful general-purpose tool for assessing PCR diagnostics assays against circulating strains.
Keywords: COVID-19; Circulating strains; Laboratory developed test; Molecular diagnostic techniques; Real-time polymerase chain reaction; SARS-CoV-2.
Crown Copyright © 2020. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors report no declarations of interest.
Figures
Similar articles
-
Multiplexing primer/probe sets for detection of SARS-CoV-2 by qRT-PCR.J Clin Virol. 2020 Aug;129:104499. doi: 10.1016/j.jcv.2020.104499. Epub 2020 Jun 8. J Clin Virol. 2020. PMID: 32535397 Free PMC article.
-
Molecular analysis of several in-house rRT-PCR protocols for SARS-CoV-2 detection in the context of genetic variability of the virus in Colombia.Infect Genet Evol. 2020 Oct;84:104390. doi: 10.1016/j.meegid.2020.104390. Epub 2020 Jun 4. Infect Genet Evol. 2020. PMID: 32505692 Free PMC article.
-
Rapid Serological Assays and SARS-CoV-2 Real-Time Polymerase Chain Reaction Assays for the Detection of SARS-CoV-2: Comparative Study.J Med Internet Res. 2020 Oct 30;22(10):e19152. doi: 10.2196/19152. J Med Internet Res. 2020. PMID: 33031048 Free PMC article.
-
Primer design for quantitative real-time PCR for the emerging Coronavirus SARS-CoV-2.Theranostics. 2020 Jun 1;10(16):7150-7162. doi: 10.7150/thno.47649. eCollection 2020. Theranostics. 2020. PMID: 32641984 Free PMC article. Review.
-
[SARS-CoV-2 and Microbiological Diagnostic Dynamics in COVID-19 Pandemic].Mikrobiyol Bul. 2020 Jul;54(3):497-509. doi: 10.5578/mb.69839. Mikrobiyol Bul. 2020. PMID: 32755524 Review. Turkish.
Cited by
-
Emerging SARS-CoV-2 variants of concern and potential intervention approaches.Crit Care. 2021 Jul 12;25(1):244. doi: 10.1186/s13054-021-03662-x. Crit Care. 2021. PMID: 34253247 Free PMC article. Review.
-
Mutations in emerging variant of concern lineages disrupt genomic sequencing of SARS-CoV-2 clinical specimens.Int J Infect Dis. 2022 Jan;114:51-54. doi: 10.1016/j.ijid.2021.10.050. Epub 2021 Oct 29. Int J Infect Dis. 2022. PMID: 34757201 Free PMC article.
-
Decoding Covid-19 with the SARS-CoV-2 Genome.Curr Genet Med Rep. 2021;9(1):1-12. doi: 10.1007/s40142-020-00197-5. Epub 2021 Jan 9. Curr Genet Med Rep. 2021. PMID: 33457109 Free PMC article. Review.
-
Misdiagnosis of SARS-CoV-2: A Critical Review of the Influence of Sampling and Clinical Detection Methods.Med Sci (Basel). 2021 May 25;9(2):36. doi: 10.3390/medsci9020036. Med Sci (Basel). 2021. PMID: 34070530 Free PMC article. Review.
-
Multiple clades of SARS-CoV-2 were introduced to Thailand during the first quarter of 2020.Microbiol Immunol. 2021 Oct;65(10):405-409. doi: 10.1111/1348-0421.12883. Epub 2021 Sep 1. Microbiol Immunol. 2021. PMID: 33835528 Free PMC article.
References
-
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.L.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(March (7798)):265–269. - PMC - PubMed
-
- World Health Organization . 2020. Coronavirus Disease (COVID-19) Situation Report 111.https://www.who.int/docs/default-source/coronaviruse/situation-reports/2... Accessed on May 1,from.
-
- World Health Organization . 2020. Molecular Assays to Diagnose COVID-19: In-house Developed Molecular Assays.https://www.who.int/docs/default-source/coronaviruse/whoinhouseassays.pd... Accessed on May 1, from.
-
- LeBlanc J.J., Gubbay J.B., Li Y., Needle R., Radons Arneson S., Marcino D., Charest H., Desnoyers G., Dust K., Fattouh R., Garceau R., German G., Hatchette T.F., Kozak R.A., Krajden M., Kuschak T., Lang A.L.S., Levett P., Mazzulli T., McDonald R., Mubareka S., Prystajecky N., Rutherford C., Smieja M., Yu Y., Zahariadis G., Zelyas N., Bastien N. COVID-19 pandemic diagnostics investigation team of the canadian public health laboratory network (CPHLN) respiratory virus working group. Real-time PCR-based SARS-CoV-2 detection in Canadian laboratories. J. Clin. Virol. 2020;128 Jul. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

