Ribosomal stalk proteins RPLP1 and RPLP2 promote biogenesis of flaviviral and cellular multi-pass transmembrane proteins
- PMID: 32890404
- PMCID: PMC7515724
- DOI: 10.1093/nar/gkaa717
Ribosomal stalk proteins RPLP1 and RPLP2 promote biogenesis of flaviviral and cellular multi-pass transmembrane proteins
Abstract
The ribosomal stalk proteins, RPLP1 and RPLP2 (RPLP1/2), which form the ancient ribosomal stalk, were discovered decades ago but their functions remain mysterious. We had previously shown that RPLP1/2 are exquisitely required for replication of dengue virus (DENV) and other mosquito-borne flaviviruses. Here, we show that RPLP1/2 function to relieve ribosome pausing within the DENV envelope coding sequence, leading to enhanced protein stability. We evaluated viral and cellular translation in RPLP1/2-depleted cells using ribosome profiling and found that ribosomes pause in the sequence coding for the N-terminus of the envelope protein, immediately downstream of sequences encoding two adjacent transmembrane domains (TMDs). We also find that RPLP1/2 depletion impacts a ribosome density for a small subset of cellular mRNAs. Importantly, the polarity of ribosomes on mRNAs encoding multiple TMDs was disproportionately affected by RPLP1/2 knockdown, implying a role for RPLP1/2 in multi-pass transmembrane protein biogenesis. These analyses of viral and host RNAs converge to implicate RPLP1/2 as functionally important for ribosomes to elongate through ORFs encoding multiple TMDs. We suggest that the effect of RPLP1/2 at TMD associated pauses is mediated by improving the efficiency of co-translational folding and subsequent protein stability.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

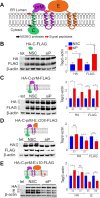
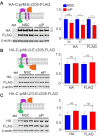

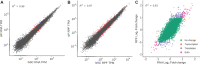


Similar articles
-
RPLP1 and RPLP2 Are Essential Flavivirus Host Factors That Promote Early Viral Protein Accumulation.J Virol. 2017 Jan 31;91(4):e01706-16. doi: 10.1128/JVI.01706-16. Print 2017 Feb 15. J Virol. 2017. PMID: 27974556 Free PMC article.
-
Senescent Cells Differentially Translate Senescence-Related mRNAs Via Ribosome Heterogeneity.J Gerontol A Biol Sci Med Sci. 2019 Jun 18;74(7):1015-1024. doi: 10.1093/gerona/gly228. J Gerontol A Biol Sci Med Sci. 2019. PMID: 30285098
-
Ribosome customization and functional diversification among P-stalk proteins regulate late poxvirus protein synthesis.Cell Rep. 2025 Jan 28;44(1):115119. doi: 10.1016/j.celrep.2024.115119. Epub 2025 Jan 8. Cell Rep. 2025. PMID: 39786991 Free PMC article.
-
The enigmatic ribosomal stalk.Q Rev Biophys. 2018 Jan;51:e12. doi: 10.1017/S0033583518000100. Q Rev Biophys. 2018. PMID: 30912488 Review.
-
Translation regulation by ribosomes: Increased complexity and expanded scope.RNA Biol. 2016 Sep;13(9):748-55. doi: 10.1080/15476286.2015.1107701. Epub 2015 Oct 29. RNA Biol. 2016. PMID: 26513496 Free PMC article. Review.
Cited by
-
RNAi-Mediated Knockdown of Acidic Ribosomal Stalk Protein P1 Arrests Egg Development in Adult Female Yellow Fever Mosquitoes, Aedes aegypti.Insects. 2024 Jan 24;15(2):84. doi: 10.3390/insects15020084. Insects. 2024. PMID: 38392504 Free PMC article.
-
Preventing translational inhibition from ribosomal protein insufficiency by a herpes simplex virus-encoded ribosome-associated protein.Proc Natl Acad Sci U S A. 2021 Nov 9;118(45):e2025546118. doi: 10.1073/pnas.2025546118. Proc Natl Acad Sci U S A. 2021. PMID: 34725147 Free PMC article.
-
Construction of a prognostic model related to copper dependence in breast cancer by single-cell sequencing analysis.Front Genet. 2022 Aug 23;13:949852. doi: 10.3389/fgene.2022.949852. eCollection 2022. Front Genet. 2022. PMID: 36082002 Free PMC article.
-
Dengue virus 3 genotype I (GI) lineage 1 (L1) isolates elicit differential cytopathic effect with syncytium formation in human glioblastoma cells (U251).Virol J. 2023 Sep 3;20(1):204. doi: 10.1186/s12985-023-02168-y. Virol J. 2023. PMID: 37661255 Free PMC article.
-
Dengue NS1 interaction with lipids alters its pathogenic effects on monocyte derived macrophages.J Biomed Sci. 2024 Sep 4;31(1):86. doi: 10.1186/s12929-024-01077-8. J Biomed Sci. 2024. PMID: 39232783 Free PMC article.
References
-
- Shi Z., Barna M.. Translating the genome in time and space: specialized ribosomes, RNA regulons, and RNA-binding proteins. Annu. Rev. Cell Dev. Biol. 2015; 31:31–54. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

