Rapid Response to Pandemic Threats: Immunogenic Epitope Detection of Pandemic Pathogens for Diagnostics and Vaccine Development Using Peptide Microarrays
- PMID: 32892628
- PMCID: PMC7640972
- DOI: 10.1021/acs.jproteome.0c00484
Rapid Response to Pandemic Threats: Immunogenic Epitope Detection of Pandemic Pathogens for Diagnostics and Vaccine Development Using Peptide Microarrays
Abstract
Emergence and re-emergence of pathogens bearing the risk of becoming a pandemic threat are on the rise. Increased travel and trade, growing population density, changes in urbanization, and climate have a critical impact on infectious disease spread. Currently, the world is confronted with the emergence of a novel coronavirus SARS-CoV-2, responsible for yet more than 800 000 deaths globally. Outbreaks caused by viruses, such as SARS-CoV-2, HIV, Ebola, influenza, and Zika, have increased over the past decade, underlining the need for a rapid development of diagnostics and vaccines. Hence, the rational identification of biomarkers for diagnostic measures on the one hand, and antigenic targets for vaccine development on the other, are of utmost importance. Peptide microarrays can display large numbers of putative target proteins translated into overlapping linear (and cyclic) peptides for a multiplexed, high-throughput antibody analysis. This enabled for example the identification of discriminant/diagnostic epitopes in Zika or influenza and mapping epitope evolution in natural infections versus vaccinations. In this review, we highlight synthesis platforms that facilitate fast and flexible generation of high-density peptide microarrays. We further outline the multifaceted applications of these peptide array platforms for the development of serological tests and vaccines to quickly encounter pandemic threats.
Keywords: array synthesis technologies; epitope mapping; infectious diseases; microarrays.
Conflict of interest statement
The authors declare the following competing financial interest(s): K.H. and L.K.W. are employees of PEPperPRINT GmbH, Germany, producing and selling peptide microarrays with a laser printer. F.F.L. is named on a patent for the production of microarrays via laser transfer. The funders had no role in the design, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish.
Figures

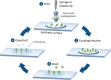


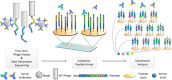
References
-
- Madhav N.; Oppenheim B.; Gallivan M.; Mulembakani P.; Rubin E.; Wolfe N.. Pandemics: Risks, Impacts, and Mitigation. In Disease Control Priorities: Improving Health and Reducing Poverty; Jamison D. T., Gelband H., Horton S., Jha P., Laxminarayan R., Mock C. N., Nugent R., Eds.; The International Bank for Reconstruction and Development/The World Bank: Washington, DC, 2017. DOI: 10.1596/978-1-4648-0527-1_ch17. - DOI - PubMed
-
- Johns Hopkins University . https://coronavirus.jhu.edu/map.html (accessed April 9, 2020).
-
- Porta M. S.; Greenland S.; Herna′n M.; Silva I. d. S.; Last J. M.; International Epidemiological Association . A Dictionary of Epidemiology, 6th ed.; Oxford University Press: Oxford, 2014; p xxxii, 343 pages.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

