The Atlas of Inflammation Resolution (AIR)
- PMID: 32893032
- PMCID: PMC7733955
- DOI: 10.1016/j.mam.2020.100894
The Atlas of Inflammation Resolution (AIR)
Abstract
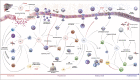
Acute inflammation is a protective reaction by the immune system in response to invading pathogens or tissue damage. Ideally, the response should be localized, self-limited, and returning to homeostasis. If not resolved, acute inflammation can result in organ pathologies leading to chronic inflammatory phenotypes. Acute inflammation and inflammation resolution are complex coordinated processes, involving a number of cell types, interacting in space and time. The biomolecular complexity and the fact that several biomedical fields are involved, make a multi- and interdisciplinary approach necessary. The Atlas of Inflammation Resolution (AIR) is a web-based resource capturing an essential part of the state-of-the-art in acute inflammation and inflammation resolution research. The AIR provides an interface for users to search thousands of interactions, arranged in inter-connected multi-layers of process diagrams, covering a wide range of clinically relevant phenotypes. By mapping experimental data onto the AIR, it can be used to elucidate drug action as well as molecular mechanisms underlying different disease phenotypes. For the visualization and exploration of information, the AIR uses the Minerva platform, which is a well-established tool for the presentation of disease maps. The molecular details of the AIR are encoded using international standards. The AIR was created as a freely accessible resource, supporting research and education in the fields of acute inflammation and inflammation resolution. The AIR connects research communities, facilitates clinical decision making, and supports research scientists in the formulation and validation of hypotheses. The AIR is accessible through https://air.bio.informatik.uni-rostock.de.
Keywords: Acute inflammation; Disease map; Inflammation resolution; Inflammatory mediators; Molecular interaction map; Molecular switches; Pro-resolving mediators; Systems biology.
Copyright © 2020 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
The AIR is built from experimentally validated information from the literature, with no information related to products of pharma companies being referred to. All authors declare that there are no competing financial interests that could undermine the objectivity, integrity and value of a publication.
Figures






References
-
- Anthony P. Robbins' pathologic basis of disease. Journal of Clinical Pathology. W.B. Saunders, Philadelphia. 1990 doi: 10.1136/jcp.43.2.176-a. fifth ed. - DOI
-
- Bara O., Day J., Djouadi S.M. Proceedings of the IEEE Conference on Decision and Control. 2013. Nonlinear state estimation for complex immune responses; pp. 3373–3378. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

