Identification of loci controlling mineral element concentration in soybean seeds
- PMID: 32894046
- PMCID: PMC7487956
- DOI: 10.1186/s12870-020-02631-w
Identification of loci controlling mineral element concentration in soybean seeds
Abstract
Background: Mineral nutrients play a crucial role in the biochemical and physiological functions of biological systems. The enhancement of seed mineral content via genetic improvement is considered as the most promising and cost-effective approach compared alternative means for meeting the dietary needs. The overall objective of this study was to perform a GWAS of mineral content (Ca, K, P and S) in seeds of a core set of 137 soybean lines that are representative of the diversity of early maturing soybeans cultivated in Canada (maturity groups 000-II).
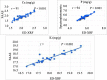
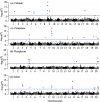
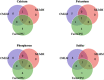
Results: This panel of 137 soybean lines was grown in five environments (in total) and the seed mineral content was measured using a portable x-ray fluorescence (XRF) spectrometer. The association analyses were carried out using three statistical models and a set of 2.2 million SNPs obtained from a combined dataset of genotyping-by-sequencing and whole-genome sequencing. Eight QTLs significantly associated with the Ca, K, P and S content were identified by at least two of the three statistical models used (in two environments) contributing each from 17 to 31% of the phenotypic variation. A strong reproducibility of the effect of seven out these eight QTLs was observed in three other environments. In total, three candidate genes were identified involved in transport and assimilation of these mineral elements.
Conclusions: There have been very few GWAS studies to identify QTLs associated with the mineral element content of soybean seeds. In addition to being new, the QTLs identified in this study and candidate genes will be useful for the genetic improvement of soybean nutritional quality through marker-assisted selection. Moreover, this study also provides details on the range of phenotypic variation encountered within the Canadian soybean germplasm.
Keywords: GWAS; Minerals; QTL; Soybean; XRF.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures




References
-
- Gibson LR, Mullen RE. Mineral concentrations in soybean seed produced under high day and night temperature. Can J Plant Sci. 2001;81:595–600. doi: 10.4141/P00-177. - DOI
-
- Kastoori Ramamurthy R, Jedlicka J, Graef GL, Waters BM. Identification of new QTLs for seed mineral, cysteine, and methionine concentrations in soybean [Glycine max (L.) Merr.] Mol Breed. 2014;34:431–445. doi: 10.1007/s11032-014-0045-z. - DOI
-
- Glass A. Physiological-mechanisms involved with genotypic differences in ion absorption and utilization. HORTSCIENCE. 1989;24:559–564.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

