Quantifying antibody kinetics and RNA detection during early-phase SARS-CoV-2 infection by time since symptom onset
- PMID: 32894217
- PMCID: PMC7508557
- DOI: 10.7554/eLife.60122
Quantifying antibody kinetics and RNA detection during early-phase SARS-CoV-2 infection by time since symptom onset
Abstract
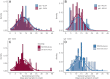
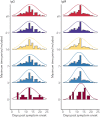
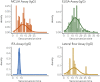
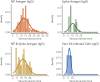
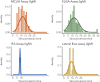
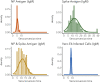
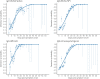
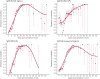
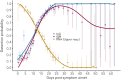
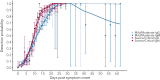
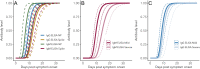
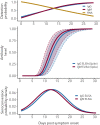
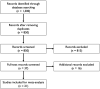
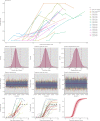
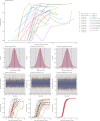
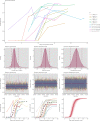
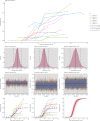
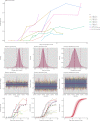
Understanding and mitigating SARS-CoV-2 transmission hinges on antibody and viral RNA data that inform exposure and shedding, but extensive variation in assays, study group demographics and laboratory protocols across published studies confounds inference of true biological patterns. Our meta-analysis leverages 3214 datapoints from 516 individuals in 21 studies to reveal that seroconversion of both IgG and IgM occurs around 12 days post-symptom onset (range 1-40), with extensive individual variation that is not significantly associated with disease severity. IgG and IgM detection probabilities increase from roughly 10% at symptom onset to 98-100% by day 22, after which IgM wanes while IgG remains reliably detectable. RNA detection probability decreases from roughly 90% to zero by day 30, and is highest in feces and lower respiratory tract samples. Our findings provide a coherent evidence base for interpreting clinical diagnostics, and for the mathematical models and serological surveys that underpin public health policies.
Keywords: COVID-19; RNA; SARS-CoV-2; antibody kinetics; detection probability; epidemiology; global health; human; meta-analysis.
© 2020, Borremans et al.
Conflict of interest statement
BB, AG, KP, SH, AM, CC, VS, JL No competing interests declared
Figures


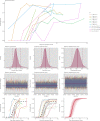
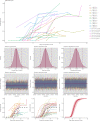
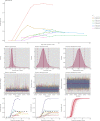
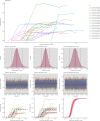
References
-
- Borremans B, Hens N, Beutels P, Leirs H, Reijniers J. Estimating time of infection using prior serological and individual information can greatly improve incidence estimation of human and wildlife infections. PLOS Computational Biology. 2016;12:e1004882. doi: 10.1371/journal.pcbi.1004882. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 707840/H2020 Marie Skłodowska-Curie Actions/International
- PREEMPT D18AC00031/Defense Advanced Research Projects Agency/International
- DEB-1557022/National Science Foundation/International
- RC‐2635/US Department of Defense Strategic Environmental Research and Development Program/International
- Cooperative Agreement W9132T1920006/Cooperative Ecosystem Studies Unit/International
LinkOut - more resources
Full Text Sources
Miscellaneous

