Cocaine-mediated circadian reprogramming in the striatum through dopamine D2R and PPARγ activation
- PMID: 32895370
- PMCID: PMC7477550
- DOI: 10.1038/s41467-020-18200-6
Cocaine-mediated circadian reprogramming in the striatum through dopamine D2R and PPARγ activation
Abstract
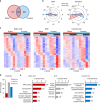
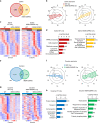
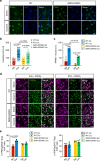
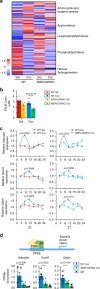
Substance abuse disorders are linked to alteration of circadian rhythms, although the molecular and neuronal pathways implicated have not been fully elucidated. Addictive drugs, such as cocaine, induce a rapid increase of dopamine levels in the brain. Here, we show that acute administration of cocaine triggers reprogramming in circadian gene expression in the striatum, an area involved in psychomotor and rewarding effects of drugs. This process involves the activation of peroxisome protein activator receptor gamma (PPARγ), a nuclear receptor involved in inflammatory responses. PPARγ reprogramming is altered in mice with cell-specific ablation of the dopamine D2 receptor (D2R) in the striatal medium spiny neurons (MSNs) (iMSN-D2RKO). Administration of a specific PPARγ agonist in iMSN-D2RKO mice elicits substantial rescue of cocaine-dependent control of circadian genes. These findings have potential implications for development of strategies to treat substance abuse disorders.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Albrecht U. Timing to perfection: the biology of central and peripheral circadian clocks. Neuron. 2012;74:246–260. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

