Polygenic architecture informs potential vulnerability to drug-induced liver injury
- PMID: 32895570
- PMCID: PMC8482794
- DOI: 10.1038/s41591-020-1023-0
Polygenic architecture informs potential vulnerability to drug-induced liver injury
Abstract
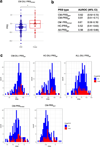
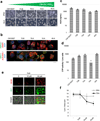
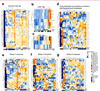
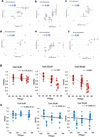
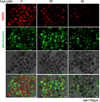
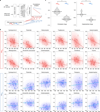
Drug-induced liver injury (DILI) is a leading cause of termination in drug development programs and removal of drugs from the market; this is partially due to the inability to identify patients who are at risk1. In this study, we developed a polygenic risk score (PRS) for DILI by aggregating effects of numerous genome-wide loci identified from previous large-scale genome-wide association studies2. The PRS predicted the susceptibility to DILI in patients treated with fasiglifam, amoxicillin-clavulanate or flucloxacillin and in primary hepatocytes and stem cell-derived organoids from multiple donors treated with over ten different drugs. Pathway analysis highlighted processes previously implicated in DILI, including unfolded protein responses and oxidative stress. In silico screening identified compounds that elicit transcriptomic signatures present in hepatocytes from individuals with elevated PRS, supporting mechanistic links and suggesting a novel screen for safety of new drug candidates. This genetic-, cellular-, organoid- and human-scale evidence underscored the polygenic architecture underlying DILI vulnerability at the level of hepatocytes, thus facilitating future mechanistic studies. Moreover, the proposed 'polygenicity-in-a-dish' strategy might potentially inform designs of safer, more efficient and robust clinical trials.
Conflict of interest statement
Competing interests
Figures








References
-
- Burant CF et al.TAK-875 versus placebo or glimepiride in type 2 diabetes mellitus: a phase 2, randomised, double-blind, placebo-controlled trial. Lancet 379, 1403–1411 (2012). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

