Effects of Cryopreservation and Thawing on Single-Cell Transcriptomes of Human T Cells
- PMID: 32895621
- PMCID: PMC7458795
- DOI: 10.4110/in.2020.20.e34
Effects of Cryopreservation and Thawing on Single-Cell Transcriptomes of Human T Cells
Abstract
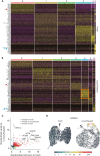
Cryopreservation and thawing of PBMCs are inevitable processes in expanding the scale of experiments in human immunology. Here, we carried out a fundamental study to investigate the detailed effects of PBMC cryopreservation and thawing on transcriptomes. We sorted Tregs from fresh and cryopreserved/thawed PBMCs from an identical donor and performed single-cell RNA-sequencing (scRNA-seq). We found that the cryopreservation and thawing process minimally affects the key molecular features of Tregs, including FOXP3. However, the cryopreserved and thawed sample had a specific cluster with up-regulation of genes for heat shock proteins. Caution may be warranted in interpreting the character of any cluster of cells with heat shock-related properties when cryopreserved and thawed samples are used for scRNA-seq.
Keywords: Cryopreservation; Single-cell analysis; T-lymphocytes, regulatory; Transcriptome.
Copyright © 2020. The Korean Association of Immunologists.
Conflict of interest statement
Conflict of Interest: The authors declare no potential conflicts of interest.
Figures


References
-
- Kim CG, Jang M, Kim Y, Leem G, Kim KH, Lee H, Kim TS, Choi SJ, Kim HD, Han JW, et al. VEGF-A drives TOX-dependent T cell exhaustion in anti-PD-1-resistant microsatellite stable colorectal cancers. Sci Immunol. 2019;4:eaay0555. - PubMed
-
- Kim HD, Song GW, Park S, Jung MK, Kim MH, Kang HJ, Yoo C, Yi K, Kim KH, Eo S, et al. Association between expression level of PD1 by tumor-infiltrating CD8(+) T cells and features of hepatocellular carcinoma. Gastroenterology. 2018;155:1936–1950.e17. - PubMed
LinkOut - more resources
Full Text Sources

