Tailoring NMR experiments for structural characterization of amorphous biological solids: A practical guide
- PMID: 32896783
- PMCID: PMC7530138
- DOI: 10.1016/j.ssnmr.2020.101686
Tailoring NMR experiments for structural characterization of amorphous biological solids: A practical guide
Abstract
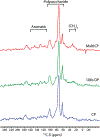
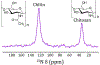
Many interesting solid-state targets for biological research do not form crystalline structures; these materials include intrinsically disordered proteins, plant biopolymer composites, cell-wall polysaccharides, and soil organic matter. The absence of aligned repeating structural elements and atomic-level rigidity presents hurdles to achieving structural elucidation and obtaining functional insights. We describe strategies for adapting several solid-state NMR methods to determine the molecular structures and compositions of these amorphous biosolids. The main spectroscopic problems in studying amorphous structures by NMR are over/under-sampling of the spin signals and spectral complexity. These problems arise in part because amorphous biosolids typically contain a mix of rigid and mobile domains, making it difficult to select a single experiment or set of acquisition conditions that fairly represents all nuclear spins in a carbon-based organic sample. These issues can be addressed by running hybrid experiments, such as using direct excitation alongside cross polarization-based methods, to develop a more holistic picture of the macromolecular system. In situations of spectral crowding or overlap, the structural elucidation strategy can be further assisted by coupling 13C spins to nuclei such as 15N, filtering out portions of the spectrum, highlighting individual moieties of interest, and adding a second or third spectral dimension to an NMR experiment in order to spread out the resonances and link them pairwise through space or through bonds. We discuss practical aspects and illustrations from the recent literature for 1D experiments that use cross or direct polarization and both homo- and heteronuclear 2D and 3D solid-state NMR experiments.
Keywords: Amorphous biosolids; Biopolymer; Correlation spectroscopy; Magic angle spinning; Solid-state NMR.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

