Development of a gene expression-based prognostic signature for IDH wild-type glioblastoma
- PMID: 32897363
- PMCID: PMC7746941
- DOI: 10.1093/neuonc/noaa157
Development of a gene expression-based prognostic signature for IDH wild-type glioblastoma
Abstract
Background: We aimed to develop a gene expression-based prognostic signature for isocitrate dehydrogenase (IDH) wild-type glioblastoma using clinical trial datasets representative of glioblastoma clinical trial populations.
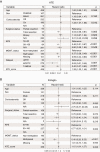
Methods: Samples were collected from newly diagnosed patients with IDH wild-type glioblastoma in the ARTE, TAMIGA, EORTC 26101 (referred to as "ATE"), AVAglio, and GLARIUS trials, or treated at UCLA. Transcriptional profiling was achieved with the NanoString gene expression platform. To identify genes prognostic for overall survival (OS), we built an elastic net penalized Cox proportional hazards regression model using the discovery ATE dataset. For validation in independent datasets (AVAglio, GLARIUS, UCLA), we combined elastic net-selected genes into a robust z-score signature (ATE score) to overcome gene expression platform differences between discovery and validation cohorts.
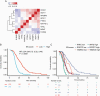
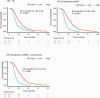
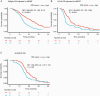
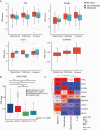
Results: NanoString data were available from 512 patients in the ATE dataset. Elastic net identified a prognostic signature of 9 genes (CHEK1, GPR17, IGF2BP3, MGMT, MTHFD1L, PTRH2, SOX11, S100A9, and TFRC). Translating weighted elastic net scores to the ATE score conserved the prognostic value of the genes. The ATE score was prognostic for OS in the ATE dataset (P < 0.0001), as expected, and in the validation cohorts (AVAglio, P < 0.0001; GLARIUS, P = 0.02; UCLA, P = 0.004). The ATE score remained prognostic following adjustment for O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation status and corticosteroid use at baseline. A positive correlation between ATE score and proneural/proliferative subtypes was observed in patients with MGMT non-methylated promoter status.
Conclusions: The ATE score showed prognostic value and may enable clinical trial stratification for IDH wild-type glioblastoma.
Keywords: NanoString; gene expression; glioblastoma; overall survival; prognostic signature.
© The Author(s) 2020. Published by Oxford University Press on behalf of the Society for Neuro-Oncology. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures

Comment in
-
Validation of the prognostic value of 9-gene ATE score for IDH wild-type glioblastoma.Neuro Oncol. 2021 Jul 1;23(7):1197-1199. doi: 10.1093/neuonc/noab058. Neuro Oncol. 2021. PMID: 33982756 Free PMC article. No abstract available.
References
-
- Louis DN, Perry A, Reifenberger G, et al. . The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. - PubMed
-
- Weller M, van den Bent M, Tonn JC, et al. ; European Association for Neuro-Oncology (EANO) Task Force on Gliomas European Association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol. 2017;18(6):e315–e329. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

