Ultra-Sensitive Serial Profiling of SARS-CoV-2 Antigens and Antibodies in Plasma to Understand Disease Progression in COVID-19 Patients with Severe Disease
- PMID: 32897389
- PMCID: PMC7499543
- DOI: 10.1093/clinchem/hvaa213
Ultra-Sensitive Serial Profiling of SARS-CoV-2 Antigens and Antibodies in Plasma to Understand Disease Progression in COVID-19 Patients with Severe Disease
Abstract
Background: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected over 21 million people worldwide since August 16, 2020. Compared to PCR and serology tests, SARS-CoV-2 antigen assays are underdeveloped, despite their potential to identify active infection and monitor disease progression.
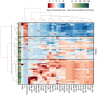
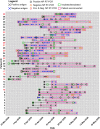
Methods: We used Single Molecule Array (Simoa) assays to quantitatively detect SARS-CoV-2 spike, S1 subunit, and nucleocapsid antigens in the plasma of patients with coronavirus disease (COVID-19). We studied plasma from 64 patients who were COVID-19 positive, 17 who were COVID-19 negative, and 34 prepandemic patients. Combined with Simoa anti-SARS-CoV-2 serological assays, we quantified changes in 31 SARS-CoV-2 biomarkers in 272 longitudinal plasma samples obtained for 39 patients with COVID-19. Data were analyzed by hierarchical clustering and were compared to longitudinal RT-PCR test results and clinical outcomes.
Results: SARS-CoV-2 S1 and N antigens were detectable in 41 out of 64 COVID-19 positive patients. In these patients, full antigen clearance in plasma was observed a mean ± 95% CI of 5 ± 1 days after seroconversion and nasopharyngeal RT-PCR tests reported positive results for 15 ± 5 days after viral-antigen clearance. Correlation between patients with high concentrations of S1 antigen and ICU admission (77%) and time to intubation (within 1 day) was statistically significant.
Conclusions: The reported SARS-CoV-2 Simoa antigen assay is the first to detect viral antigens in the plasma of patients who were COVID-19 positive to date. These data show that SARS-CoV-2 viral antigens in the blood are associated with disease progression, such as respiratory failure, in COVID-19 cases with severe disease.
Keywords: SARS-CoV-2; longitudinal plasma samples; serological; single molecule arrays; viral antigen.
© American Association for Clinical Chemistry 2020. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures



References
-
- World Health Organization. Coronavirus disease (COVID-19) Situation Report 178. 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/2... (Accessed August 2020).
-
- Sethuraman N, Jeremiah SS, Ryo A.. Interpreting diagnostic tests for SARS-CoV-2. JAMA 2020;323:2249–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

