Ultra-high field (10.5 T) resting state fMRI in the macaque
- PMID: 32898683
- PMCID: PMC7745777
- DOI: 10.1016/j.neuroimage.2020.117349
Ultra-high field (10.5 T) resting state fMRI in the macaque
Abstract
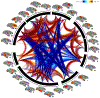
Resting state functional connectivity refers to the temporal correlations between spontaneous hemodynamic signals obtained using functional magnetic resonance imaging. This technique has demonstrated that the structure and dynamics of identifiable networks are altered in psychiatric and neurological disease states. Thus, resting state network organizations can be used as a diagnostic, or prognostic recovery indicator. However, much about the physiological basis of this technique is unknown. Thus, providing a translational bridge to an optimal animal model, the macaque, in which invasive circuit manipulations are possible, is of utmost importance. Current approaches to resting state measurements in macaques face unique challenges associated with signal-to-noise, the need for contrast agents limiting translatability, and within-subject designs. These limitations can, in principle, be overcome through ultra-high magnetic fields. However, imaging at magnetic fields above 7T has yet to be adapted for fMRI in macaques. Here, we demonstrate that the combination of high channel count transmitter and receiver arrays, optimized pulse sequences, and careful anesthesia regimens, allows for detailed single-subject resting state analysis at high resolutions using a 10.5 Tesla scanner. In this study, we uncover thirty spatially detailed resting state components that are highly robust across individual macaques and closely resemble the quality and findings of connectomes from large human datasets. This detailed map of the rsfMRI 'macaque connectome' will be the basis for future neurobiological circuit manipulation work, providing valuable biological insights into human connectomics.
Keywords: Functional MRI (fMRI); Functional connectivity; Resting-state; Rhesus macaque; Spontaneous activity.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
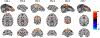





References
-
- Autio JA, Glasser MF, Ose T, Donahue CJ, Bastiani M, Ohno M, Kawabata Y, Urushibata Y, Murata K, Nishigori K, Yamaguchi M, Hori Y, Yoshida A, Go Y, Coalson TS, Jbabdi S, Sotiropoulos SN, Kennedy H, Smith S, Essen DCV, Hayashi T, 2020. Towards HCP-Style macaque connectomes: 24-Channel 3T multi-array coil, MRI sequences and preprocessing. Neuroimage 215, 116800. doi: 10.1016/j.neuroimage.2020.116800. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

