Regulation of mTORC2 Signaling
- PMID: 32899613
- PMCID: PMC7564249
- DOI: 10.3390/genes11091045
Regulation of mTORC2 Signaling
Abstract
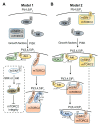
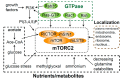
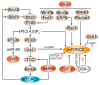
Mammalian target of rapamycin (mTOR), a serine/threonine protein kinase and a master regulator of cell growth and metabolism, forms two structurally and functionally distinct complexes, mTOR complex 1 (mTORC1) and mTORC2. While mTORC1 signaling is well characterized, mTORC2 is relatively poorly understood. mTORC2 appears to exist in functionally distinct pools, but few mTORC2 effectors/substrates have been identified. Here, we review recent advances in our understanding of mTORC2 signaling, with particular emphasis on factors that control mTORC2 activity.
Keywords: Akt; mTOR; mTORC2 signaling; signaling crosstalk.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
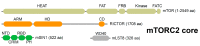
References
-
- Dos D.S., Ali S.M., Kim D.H., Guertin D.A., Latek R.R., Erdjument-Bromage H., Tempst P., Sabatini D.M. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 2004;14:1296–1302. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

