How Do mAbs Make Use of Complement to Kill Cancer Cells? The Role of Ca2
- PMID: 32899722
- PMCID: PMC7551823
- DOI: 10.3390/antib9030045
How Do mAbs Make Use of Complement to Kill Cancer Cells? The Role of Ca2
Abstract
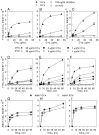
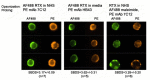
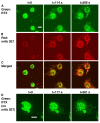
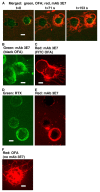
We examined the kinetics and mechanisms by which monoclonal antibodies (mAbs) utilize complement to rapidly kill targeted cancer cells. Based on results from flow cytometry, confocal microscopy and high-resolution digital imaging experiments, the general patterns which have emerged reveal cytotoxic activities mediated by substantial and lethal Ca2+ fluxes. The Ca2+ fluxes are common to the reported pathways that have been utilized by other toxins in killing nucleated cells. These reactions terminate in very high levels of cell killing, and based on these considerations, we suggest additional strategies to further enhance mAb-based targeting of cancer with complement.
Keywords: Ca2+; complement; fluorescence microscopy; therapeutic monoclonal antibodies (mAbs).
Conflict of interest statement
RPT and MAL have licensed mAbs 3E7 and 7C12 for commercial use through the University of Virginia Licensing and Ventures Group.
Figures

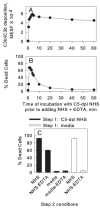
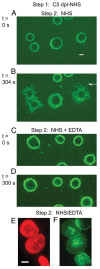


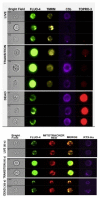
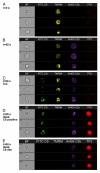


References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

