Genome-Wide Characterization, Evolution, and Expression Analysis of the Leucine-Rich Repeat Receptor-Like Protein Kinase (LRR-RLK) Gene Family in Medicago truncatula
- PMID: 32899802
- PMCID: PMC7555646
- DOI: 10.3390/life10090176
Genome-Wide Characterization, Evolution, and Expression Analysis of the Leucine-Rich Repeat Receptor-Like Protein Kinase (LRR-RLK) Gene Family in Medicago truncatula
Abstract
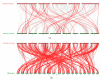
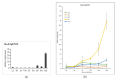
Leucine-rich repeat receptor-like kinases (LRR-RLKs) constitute the largest subfamily of receptor-like kinases (RLKs) in plants. They play roles in plant growth and developmental and physiological processes, but less is known about the functions of LRR-RLKs in Medicago truncatula. Our genome-wide analysis revealed 329 LRR-RLK genes in the M.truncatula genome. Phylogenetic and classification analysis suggested that these genes could be classified into 15 groups and 24 subgroups. A total of 321 genes were mapped onto all chromosomes, and 23 tandem duplications (TDs) involving 56 genes were distributed on each chromosome except 4. Twenty-seven M.truncatula LRR-RLK segmental duplication gene pairs were colinearly related. The exon/intron organization, motif composition and arrangements were relatively conserved among members of the same groups or subgroups. Using publicly available RNAseq data and quantitative real-time polymerase chain reaction (qRT-PCR), expression profiling suggested that LRR-RLKs were differentially expressed among different tissues, while some were expressed specifically in the roots and nodules. The expression of LRR-RLKs in A17 and 4 nodule mutants under rhizobial infection showed that 36 LRR-RKLs were highly upregulated in the sickle (skl) mutant [an ethylene (ET)-insensitive, Nod factor-hypersensitive mutant] after 12 h of rhizobium inoculation. Among these LRR-RLKs, six genes were also expressed specifically in the roots and nodules, which might be specific to the Nod factor and involved in autoregulation of the nodulation signal. Our results provide information on the LRR-RLK gene family in M. truncatula and serve as a guide for functional research of the LRR-RLKs.
Keywords: M. truncatula; evolutionary analysis; expression profiling; leucine-rich repeat receptor-like kinase (LRR-RLKs); phylogenetic analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
Grants and funding
- No. 31100871 and No.31672191/the National Natural Science Foundation of China
- No.2015ZCQ-YL-03/Central University's basic research business expenses
- No.YX2015-16/Central University's basic research business fee special fund
- No.2019XKJS0323/Beijing Forestry University to build a world-class discipline and characteristics development guidance Special funds
LinkOut - more resources
Full Text Sources

