Isolation and Cultivation of a New Isolate of BTV-25 and Presumptive Evidence for a Potential Persistent Infection in Healthy Goats
- PMID: 32899808
- PMCID: PMC7552037
- DOI: 10.3390/v12090983
Isolation and Cultivation of a New Isolate of BTV-25 and Presumptive Evidence for a Potential Persistent Infection in Healthy Goats
Abstract
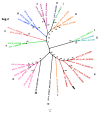
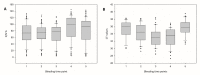
Recently, several so-called "atypical" Bluetongue virus (BTV) serotypes were discovered, including BTV-25 (Toggenburg virus), in Switzerland. Most "atypical" BTV were identified in small ruminants without clinical signs. In 2018, two goats from a holding in Germany tested positive for BTV-25 genome by RT-qPCR prior to export. After experimental inoculation of the two goats with the BTV-25 positive field blood samples for generation of reference materials, viremia could be observed in one animal. For the first time, the BTV-25-related virus was isolated in cell culture from EDTA-blood and the full genome of isolate "BTV-25-GER2018" could be generated. BTV-25-GER2018 was only incompletely neutralized by ELISA-positive sera. We could monitor the BTV-25 occurrence in the respective affected goat flock of approximately 120 goats over several years. EDTA blood samples were screened with RT-qPCR using a newly developed BTV-25 specific assay. For serological surveillance, serum samples were screened using a commercial cELISA. BTV-25-GER2018 was detected over 4.5 years in the goat flock with intermittent PCR-positivity in some animals, and with or without concomitantly detected antibodies since 2015. We could demonstrate the viral persistence of BTV-25-GER2018 in goats for up to 4.5 years, and the first BTV-25 isolate is now available for further characterization.
Keywords: BTV; Bluetongue virus; atypical BTV; goats; persistent infection; serotype 25.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures




References
-
- Mertens P.P., Diprose J., Maan S., Singh K.P., Attoui H., Samuel A.R. Bluetongue virus replication, molecular and structural biology. Vet. Ital. 2004;40:426–437. - PubMed
-
- Owen N.C., Du Toit R.M., Howell P.G. Bluetongue in cattle: Typing of viruses isolated from cattle exposed to natural infections. Onderstepoort J. Vet. Res. 1965;32:3–6. - PubMed
-
- Breard E., Schulz C., Sailleau C., Bernelin-Cottet C., Viarouge C., Vitour D., Guillaume B., Caignard G., Gorlier A., Attoui H., et al. Bluetongue virus serotype 27: Experimental infection of goats, sheep and cattle with three BTV-27 variants reveal atypical characteristics and likely direct contact transmission BTV-27 between goats. Transbound Emerg. Dis. 2018;65:e251–e263. doi: 10.1111/tbed.12780. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

