Sigma 54-Regulated Transcription Is Associated with Membrane Reorganization and Type III Secretion Effectors during Conversion to Infectious Forms of Chlamydia trachomatis
- PMID: 32900805
- PMCID: PMC7482065
- DOI: 10.1128/mBio.01725-20
Sigma 54-Regulated Transcription Is Associated with Membrane Reorganization and Type III Secretion Effectors during Conversion to Infectious Forms of Chlamydia trachomatis
Abstract
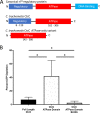
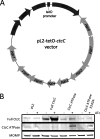
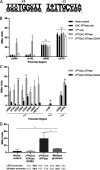
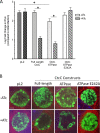
Chlamydia bacteria are obligate intracellular organisms with a phylum-defining biphasic developmental cycle that is intrinsically linked to its ability to cause disease. The progression of the chlamydial developmental cycle is regulated by the temporal expression of genes predominantly controlled by RNA polymerase sigma (σ) factors. Sigma 54 (σ54) is one of three sigma factors encoded by Chlamydia for which the role and regulon are unknown. CtcC is part of a two-component signal transduction system that is requisite for σ54 transcriptional activation. CtcC activation of σ54 requires phosphorylation, which relieves inhibition by the CtcC regulatory domain and enables ATP hydrolysis by the ATPase domain. Prior studies with CtcC homologs in other organisms have shown that expression of the ATPase domain alone can activate σ54 transcription. Biochemical analysis of CtcC ATPase domain supported the idea of ATP hydrolysis occurring in the absence of the regulatory domain, as well as the presence of an active-site residue essential for ATPase activity (E242). Using recently developed genetic approaches in Chlamydia to induce expression of the CtcC ATPase domain, a transcriptional profile was determined that is expected to reflect the σ54 regulon. Computational evaluation revealed that the majority of the differentially expressed genes were preceded by highly conserved σ54 promoter elements. Reporter gene analyses using these putative σ54 promoters reinforced the accuracy of the model of the proposed regulon. Investigation of the gene products included in this regulon supports the idea that σ54 controls expression of genes that are critical for conversion of Chlamydia from replicative reticulate bodies into infectious elementary bodies.IMPORTANCE The factors that control the growth and infectious processes for Chlamydia are still poorly understood. This study used recently developed genetic tools to determine the regulon for one of the key transcription factors encoded by Chlamydia, sigma 54. Surrogate and computational analyses provide additional support for the hypothesis that sigma 54 plays a key role in controlling the expression of many components critical to converting and enabling the infectious capability of Chlamydia These components include those that remodel the membrane for the extracellular environment and incorporation of an arsenal of type III secretion effectors in preparation for infecting new cells.
Keywords: Chlamydia trachomatis; gene regulation; regulon; sigma factors.
Copyright © 2020 Soules et al.
Figures
Similar articles
-
Type III Secretion in Chlamydia.Microbiol Mol Biol Rev. 2023 Sep 26;87(3):e0003423. doi: 10.1128/mmbr.00034-23. Epub 2023 Jun 26. Microbiol Mol Biol Rev. 2023. PMID: 37358451 Free PMC article. Review.
-
Identification of the alternative sigma factor regulons of Chlamydia trachomatis using multiplexed CRISPR interference.mSphere. 2023 Oct 24;8(5):e0039123. doi: 10.1128/msphere.00391-23. Epub 2023 Sep 25. mSphere. 2023. PMID: 37747235 Free PMC article.
-
Identification of a GrgA-Euo-HrcA Transcriptional Regulatory Network in Chlamydia.mSystems. 2021 Aug 31;6(4):e0073821. doi: 10.1128/mSystems.00738-21. Epub 2021 Aug 3. mSystems. 2021. PMID: 34342542 Free PMC article.
-
Role for GrgA in Regulation of σ28-Dependent Transcription in the Obligate Intracellular Bacterial Pathogen Chlamydia trachomatis.J Bacteriol. 2018 Sep 24;200(20):e00298-18. doi: 10.1128/JB.00298-18. Print 2018 Oct 15. J Bacteriol. 2018. PMID: 30061357 Free PMC article.
-
The Regulatory Functions of σ54 Factor in Phytopathogenic Bacteria.Int J Mol Sci. 2021 Nov 24;22(23):12692. doi: 10.3390/ijms222312692. Int J Mol Sci. 2021. PMID: 34884502 Free PMC article. Review.
Cited by
-
In-cell western assay as a high-throughput approach for Chlamydia trachomatis quantification and susceptibility testing to antimicrobials.PLoS One. 2021 May 11;16(5):e0251075. doi: 10.1371/journal.pone.0251075. eCollection 2021. PLoS One. 2021. PMID: 33974662 Free PMC article.
-
Molecular pathogenesis of Chlamydia trachomatis.Front Cell Infect Microbiol. 2023 Oct 18;13:1281823. doi: 10.3389/fcimb.2023.1281823. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37920447 Free PMC article. Review.
-
Flip the switch: the role of FleQ in modulating the transition between the free-living and sessile mode of growth in Pseudomonas aeruginosa.J Bacteriol. 2024 Mar 21;206(3):e0036523. doi: 10.1128/jb.00365-23. Epub 2024 Mar 4. J Bacteriol. 2024. PMID: 38436566 Free PMC article. Review.
-
Type III Secretion in Chlamydia.Microbiol Mol Biol Rev. 2023 Sep 26;87(3):e0003423. doi: 10.1128/mmbr.00034-23. Epub 2023 Jun 26. Microbiol Mol Biol Rev. 2023. PMID: 37358451 Free PMC article. Review.
-
Characterization of the RpoN regulon reveals the regulation of motility, T6SS2 and metabolism in Vibrio parahaemolyticus.Front Microbiol. 2022 Dec 22;13:1025960. doi: 10.3389/fmicb.2022.1025960. eCollection 2022. Front Microbiol. 2022. PMID: 36620062 Free PMC article.
References
-
- Niehus E, Gressmann H, Ye F, Schlapbach R, Dehio M, Dehio C, Stack A, Meyer TF, Suerbaum S, Josenhans C. 2004. Genome-wide analysis of transcriptional hierarchy and feedback regulation in the flagellar system of Helicobacter pylori. Mol Microbiol 52:947–961. doi:10.1111/j.1365-2958.2004.04006.x. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
