Genome-wide variation and transcriptional changes in diverse developmental processes underlie the rapid evolution of seasonal adaptation
- PMID: 32900926
- PMCID: PMC7519392
- DOI: 10.1073/pnas.2002357117
Genome-wide variation and transcriptional changes in diverse developmental processes underlie the rapid evolution of seasonal adaptation
Abstract
Many organisms enter a dormant state in their life cycle to deal with predictable changes in environments over the course of a year. The timing of dormancy is therefore a key seasonal adaptation, and it evolves rapidly with changing environments. We tested the hypothesis that differences in the timing of seasonal activity are driven by differences in the rate of development during diapause in Rhagoletis pomonella, a fly specialized to feed on fruits of seasonally limited host plants. Transcriptomes from the central nervous system across a time series during diapause show consistent and progressive changes in transcripts participating in diverse developmental processes, despite a lack of gross morphological change. Moreover, population genomic analyses suggested that many genes of small effect enriched in developmental functional categories underlie variation in dormancy timing and overlap with gene sets associated with development rate in Drosophila melanogaster Our transcriptional data also suggested that a recent evolutionary shift from a seasonally late to a seasonally early host plant drove more rapid development during diapause in the early fly population. Moreover, genetic variants that diverged during the evolutionary shift were also enriched in putative cis regulatory regions of genes differentially expressed during diapause development. Overall, our data suggest polygenic variation in the rate of developmental progression during diapause contributes to the evolution of seasonality in R. pomonella We further discuss patterns that suggest hourglass-like developmental divergence early and late in diapause development and an important role for hub genes in the evolution of transcriptional divergence.
Keywords: adaptation; development; diapause; ecological genomics; phenology.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures


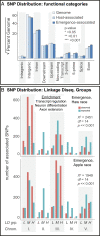
Similar articles
-
A rapidly evolved shift in life-history timing during ecological speciation is driven by the transition between developmental phases.J Evol Biol. 2020 Oct;33(10):1371-1386. doi: 10.1111/jeb.13676. Epub 2020 Aug 5. J Evol Biol. 2020. PMID: 32649797
-
Divergence of the diapause transcriptome in apple maggot flies: winter regulation and post-winter transcriptional repression.J Exp Biol. 2016 Sep 1;219(Pt 17):2613-22. doi: 10.1242/jeb.140566. Epub 2016 Jun 16. J Exp Biol. 2016. PMID: 27312473
-
Developmental trajectories of gene expression reveal candidates for diapause termination: a key life-history transition in the apple maggot fly Rhagoletis pomonella.J Exp Biol. 2011 Dec 1;214(Pt 23):3948-59. doi: 10.1242/jeb.061085. J Exp Biol. 2011. PMID: 22071185
-
Animal-Microbe Interactions in the Context of Diapause.Biol Bull. 2019 Oct;237(2):180-191. doi: 10.1086/706078. Epub 2019 Oct 23. Biol Bull. 2019. PMID: 31714855 Review.
-
What makes a fly enter diapause?Fly (Austin). 2007 Nov-Dec;1(6):307-10. doi: 10.4161/fly.5532. Epub 2007 Nov 7. Fly (Austin). 2007. PMID: 18820432 Review.
Cited by
-
QTL identification and characterization of the recombination landscape of the mountain pine beetle (Dendroctonus ponderosae).G3 (Bethesda). 2025 Jul 9;15(7):jkaf101. doi: 10.1093/g3journal/jkaf101. G3 (Bethesda). 2025. PMID: 40333319 Free PMC article.
-
Rapid brain development and reduced neuromodulator titres correlate with host shifts in Rhagoletis pomonella.R Soc Open Sci. 2022 Sep 7;9(9):220962. doi: 10.1098/rsos.220962. eCollection 2022 Sep. R Soc Open Sci. 2022. PMID: 36117862 Free PMC article.
-
The Build-Up of Population Genetic Divergence along the Speciation Continuum during a Recent Adaptive Radiation of Rhagoletis Flies.Genes (Basel). 2022 Jan 30;13(2):275. doi: 10.3390/genes13020275. Genes (Basel). 2022. PMID: 35205320 Free PMC article.
-
Chromosome-level genome assembly of an important wolfberry fruit fly (Neoceratitis asiatica Becker).Sci Data. 2023 Oct 4;10(1):675. doi: 10.1038/s41597-023-02601-5. Sci Data. 2023. PMID: 37794161 Free PMC article.
-
Diapause-Linked Gene Expression Pattern and Related Candidate Duplicated Genes of the Mountain Butterfly Parnassius glacialis (Lepidoptera: Papilionidae) Revealed by Comprehensive Transcriptome Profiling.Int J Mol Sci. 2023 Mar 14;24(6):5577. doi: 10.3390/ijms24065577. Int J Mol Sci. 2023. PMID: 36982649 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

