Lifestyle adaptations of Rhizobium from rhizosphere to symbiosis
- PMID: 32900931
- PMCID: PMC7519234
- DOI: 10.1073/pnas.2009094117
Lifestyle adaptations of Rhizobium from rhizosphere to symbiosis
Abstract
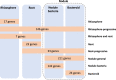
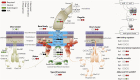
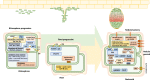
By analyzing successive lifestyle stages of a model Rhizobium-legume symbiosis using mariner-based transposon insertion sequencing (INSeq), we have defined the genes required for rhizosphere growth, root colonization, bacterial infection, N2-fixing bacteroids, and release from legume (pea) nodules. While only 27 genes are annotated as nif and fix in Rhizobium leguminosarum, we show 603 genetic regions (593 genes, 5 transfer RNAs, and 5 RNA features) are required for the competitive ability to nodulate pea and fix N2 Of these, 146 are common to rhizosphere growth through to bacteroids. This large number of genes, defined as rhizosphere-progressive, highlights how critical successful competition in the rhizosphere is to subsequent infection and nodulation. As expected, there is also a large group (211) specific for nodule bacteria and bacteroid function. Nodule infection and bacteroid formation require genes for motility, cell envelope restructuring, nodulation signaling, N2 fixation, and metabolic adaptation. Metabolic adaptation includes urea, erythritol and aldehyde metabolism, glycogen synthesis, dicarboxylate metabolism, and glutamine synthesis (GlnII). There are 17 separate lifestyle adaptations specific to rhizosphere growth and 23 to root colonization, distinct from infection and nodule formation. These results dramatically highlight the importance of competition at multiple stages of a Rhizobium-legume symbiosis.
Keywords: N2 fixation; Rhizobium; legume; nodulation; rhizosphere.
Conflict of interest statement
The authors declare no competing interest.
Figures


Similar articles
-
Rhizobium determinants of rhizosphere persistence and root colonization.ISME J. 2024 Jan 8;18(1):wrae072. doi: 10.1093/ismejo/wrae072. ISME J. 2024. PMID: 38690786 Free PMC article.
-
Optimizing Rhizobium-legume symbioses by simultaneous measurement of rhizobial competitiveness and N2 fixation in nodules.Proc Natl Acad Sci U S A. 2020 May 5;117(18):9822-9831. doi: 10.1073/pnas.1921225117. Epub 2020 Apr 21. Proc Natl Acad Sci U S A. 2020. PMID: 32317381 Free PMC article.
-
Identifying abnormalities in symbiotic development between Trifolium spp. and Rhizobium leguminosarum bv. trifolii leading to sub-optimal and ineffective nodule phenotypes.Ann Bot. 2012 Dec;110(8):1559-72. doi: 10.1093/aob/mcs206. Epub 2012 Sep 17. Ann Bot. 2012. PMID: 22989463 Free PMC article.
-
Evolution of NIN and NIN-like Genes in Relation to Nodule Symbiosis.Genes (Basel). 2020 Jul 11;11(7):777. doi: 10.3390/genes11070777. Genes (Basel). 2020. PMID: 32664480 Free PMC article. Review.
-
To keep or not to keep: mRNA stability and translatability in root nodule symbiosis.Curr Opin Plant Biol. 2020 Aug;56:109-117. doi: 10.1016/j.pbi.2020.04.012. Epub 2020 Jun 20. Curr Opin Plant Biol. 2020. PMID: 32569975 Review.
Cited by
-
Novel lineages of single-stranded DNA phages that coevolved with the symbiotic bacteria Rhizobium.Front Microbiol. 2022 Sep 13;13:990394. doi: 10.3389/fmicb.2022.990394. eCollection 2022. Front Microbiol. 2022. PMID: 36177468 Free PMC article.
-
How It All Begins: Bacterial Factors Mediating the Colonization of Invertebrate Hosts by Beneficial Symbionts.Microbiol Mol Biol Rev. 2022 Dec 21;86(4):e0012621. doi: 10.1128/mmbr.00126-21. Epub 2022 Oct 27. Microbiol Mol Biol Rev. 2022. PMID: 36301103 Free PMC article. Review.
-
Genomic Diversity of Pigeon Pea (Cajanus cajan L. Millsp.) Endosymbionts in India and Selection of Potential Strains for Use as Agricultural Inoculants.Front Plant Sci. 2021 Sep 7;12:680981. doi: 10.3389/fpls.2021.680981. eCollection 2021. Front Plant Sci. 2021. PMID: 34557206 Free PMC article.
-
How Rhizobia Adapt to the Nodule Environment.J Bacteriol. 2021 May 20;203(12):e0053920. doi: 10.1128/JB.00539-20. Epub 2021 Feb 1. J Bacteriol. 2021. PMID: 33526611 Free PMC article.
-
Unraveling Burkholderia cenocepacia H111 fitness determinants using two animal models.mSystems. 2025 Apr 22;10(4):e0135424. doi: 10.1128/msystems.01354-24. Epub 2025 Mar 19. mSystems. 2025. PMID: 40105327 Free PMC article.
References
-
- Galloway J. N. et al. ., Transformation of the nitrogen cycle: Recent trends, questions, and potential solutions. Science 320, 889–892 (2008). - PubMed
-
- Poole P., Allaway D., Carbon and nitrogen metabolism in Rhizobium. Adv. Microb. Physiol. 43, 117–163 (2000). - PubMed
-
- Hardy R. W. F., Havelka U. D., Nitrogen fixation research: A key to world food? Science 188, 633–643 (1975). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

