HDAC3 deacetylates the DNA mismatch repair factor MutSβ to stimulate triplet repeat expansions
- PMID: 32900932
- PMCID: PMC7519323
- DOI: 10.1073/pnas.2013223117
HDAC3 deacetylates the DNA mismatch repair factor MutSβ to stimulate triplet repeat expansions
Abstract
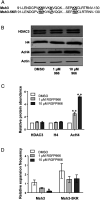
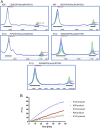
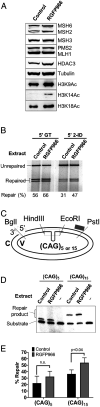
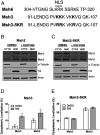
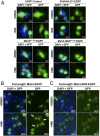
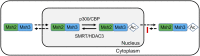
Trinucleotide repeat (TNR) expansions cause nearly 20 severe human neurological diseases which are currently untreatable. For some of these diseases, ongoing somatic expansions accelerate disease progression and may influence age of onset. This new knowledge emphasizes the importance of understanding the protein factors that drive expansions. Recent genetic evidence indicates that the mismatch repair factor MutSβ (Msh2-Msh3 complex) and the histone deacetylase HDAC3 function in the same pathway to drive triplet repeat expansions. Here we tested the hypothesis that HDAC3 deacetylates MutSβ and thereby activates it to drive expansions. The HDAC3-selective inhibitor RGFP966 was used to examine its biological and biochemical consequences in human tissue culture cells. HDAC3 inhibition efficiently suppresses repeat expansion without impeding canonical mismatch repair activity. Five key lysine residues in Msh3 are direct targets of HDAC3 deacetylation. In cells expressing Msh3 in which these lysine residues are mutated to arginine, the inhibitory effect of RGFP966 on expansions is largely bypassed, consistent with the direct deacetylation hypothesis. RGFP966 treatment does not alter MutSβ subunit abundance or complex formation but does partially control its subcellular localization. Deacetylation sites in Msh3 overlap a nuclear localization signal, and we show that localization of MutSβ is partially dependent on HDAC3 activity. Together, these results indicate that MutSβ is a key target of HDAC3 deacetylation and provide insights into an innovative regulatory mechanism for triplet repeat expansions. The results suggest expansion activity may be druggable and support HDAC3-selective inhibition as an attractive therapy in some triplet repeat expansion diseases.
Keywords: histone deacetylase 3; mismatch repair; triplet repeat expansion.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures
Similar articles
-
A selective inhibitor of histone deacetylase 3 prevents cognitive deficits and suppresses striatal CAG repeat expansions in Huntington's disease mice.Sci Rep. 2017 Jul 20;7(1):6082. doi: 10.1038/s41598-017-05125-2. Sci Rep. 2017. PMID: 28729730 Free PMC article.
-
MutSβ and histone deacetylase complexes promote expansions of trinucleotide repeats in human cells.Nucleic Acids Res. 2012 Nov 1;40(20):10324-33. doi: 10.1093/nar/gks810. Epub 2012 Aug 31. Nucleic Acids Res. 2012. PMID: 22941650 Free PMC article.
-
MutSβ abundance and Msh3 ATP hydrolysis activity are important drivers of CTG•CAG repeat expansions.Nucleic Acids Res. 2017 Sep 29;45(17):10068-10078. doi: 10.1093/nar/gkx650. Nucleic Acids Res. 2017. PMID: 28973443 Free PMC article.
-
Disease-associated repeat instability and mismatch repair.DNA Repair (Amst). 2016 Feb;38:117-126. doi: 10.1016/j.dnarep.2015.11.008. Epub 2015 Dec 12. DNA Repair (Amst). 2016. PMID: 26774442 Review.
-
DNA triplet repeat expansion and mismatch repair.Annu Rev Biochem. 2015;84:199-226. doi: 10.1146/annurev-biochem-060614-034010. Epub 2015 Jan 2. Annu Rev Biochem. 2015. PMID: 25580529 Free PMC article. Review.
Cited by
-
Overview of the Complex Relationship between Epigenetics Markers, CTG Repeat Instability and Symptoms in Myotonic Dystrophy Type 1.Int J Mol Sci. 2022 Mar 23;23(7):3477. doi: 10.3390/ijms23073477. Int J Mol Sci. 2022. PMID: 35408837 Free PMC article. Review.
-
Factors influencing reduced penetrance and variable expressivity in X-linked dystonia-parkinsonism.Med Genet. 2022 Aug 12;34(2):97-102. doi: 10.1515/medgen-2022-2135. eCollection 2022 Jun. Med Genet. 2022. PMID: 38835911 Free PMC article.
-
New developments in Huntington's disease and other triplet repeat diseases: DNA repair turns to the dark side.Neuronal Signal. 2020 Nov 16;4(4):NS20200010. doi: 10.1042/NS20200010. eCollection 2020 Dec. Neuronal Signal. 2020. PMID: 33224521 Free PMC article. Review.
-
Identifying genetic modifiers of age-associated penetrance in X-linked dystonia-parkinsonism.Nat Commun. 2021 May 28;12(1):3216. doi: 10.1038/s41467-021-23491-4. Nat Commun. 2021. PMID: 34050153 Free PMC article.
-
Modifiers of CAG/CTG Repeat Instability: Insights from Mammalian Models.J Huntingtons Dis. 2021;10(1):123-148. doi: 10.3233/JHD-200426. J Huntingtons Dis. 2021. PMID: 33579861 Free PMC article. Review.
References
-
- The Huntington’s Disease Collaborative Research Group , A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 72, 971–983 (1993). - PubMed
-
- Brook J. D. et al. ., Molecular basis of myotonic dystrophy: Expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell 68, 799–808 (1992). - PubMed
-
- Fu Y.-H. et al. ., An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science 255, 1256–1258 (1992). - PubMed
-
- Mahadevan M. et al. ., Myotonic dystrophy mutation: An unstable CTG repeat in the 3′ untranslated region of the gene. Science 255, 1253–1255 (1992). - PubMed
-
- Kremer E. J. et al. ., Mapping of DNA instability at the fragile X to a trinucleotide repeat sequence p(CCG)n. Science 252, 1711–1714 (1991). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

