SARS-CoV-2 detection using isothermal amplification and a rapid, inexpensive protocol for sample inactivation and purification
- PMID: 32900935
- PMCID: PMC7533677
- DOI: 10.1073/pnas.2011221117
SARS-CoV-2 detection using isothermal amplification and a rapid, inexpensive protocol for sample inactivation and purification
Abstract
The current severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has had an enormous impact on society worldwide, threatening the lives and livelihoods of many. The effects will continue to grow and worsen if economies begin to open without the proper precautions, including expanded diagnostic capabilities. To address this need for increased testing, we have developed a sensitive reverse-transcription loop-mediated isothermal amplification (RT-LAMP) assay compatible with current reagents, which utilizes a colorimetric readout in as little as 30 min. A rapid inactivation protocol capable of inactivating virions, as well as endogenous nucleases, was optimized to increase sensitivity and sample stability. This protocol, combined with the RT-LAMP assay, has a sensitivity of at least 50 viral RNA copies per microliter in a sample. To further increase the sensitivity, a purification protocol compatible with this inactivation method was developed. The inactivation and purification protocol, combined with the RT-LAMP assay, brings the sensitivity to at least 1 viral RNA copy per microliter in a sample. This simple inactivation and purification pipeline is inexpensive and compatible with other downstream RNA detection platforms and uses readily available reagents. It should increase the availability of SARS-CoV-2 testing as well as expand the settings in which this testing can be performed.
Keywords: RT-LAMP; SARS-CoV-2; diagnostic.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures
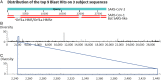

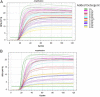





References
-
- Zhang Y., et al. , Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric LAMP. MedRxiv:2020.02.26.20028373 (29 February 2020).
-
- Nagamine K., Hase T., Notomi T., Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 16, 223–229 (2002). - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

