p53 drives a transcriptional program that elicits a non-cell-autonomous response and alters cell state in vivo
- PMID: 32900967
- PMCID: PMC7519296
- DOI: 10.1073/pnas.2008474117
p53 drives a transcriptional program that elicits a non-cell-autonomous response and alters cell state in vivo
Abstract
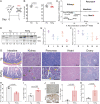
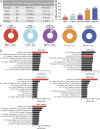
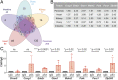
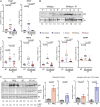
Cell stress and DNA damage activate the tumor suppressor p53, triggering transcriptional activation of a myriad of target genes. The molecular, morphological, and physiological consequences of this activation remain poorly understood in vivo. We activated a p53 transcriptional program in mice by deletion of Mdm2, a gene that encodes the major p53 inhibitor. By overlaying tissue-specific RNA-sequencing data from pancreas, small intestine, ovary, kidney, and heart with existing p53 chromatin immunoprecipitation (ChIP) sequencing, we identified a large repertoire of tissue-specific p53 genes and a common p53 transcriptional signature of seven genes, which included Mdm2 but not p21 Global p53 activation caused a metaplastic phenotype in the pancreas that was missing in mice with acinar-specific p53 activation, suggesting non-cell-autonomous effects. The p53 cellular response at single-cell resolution in the intestine altered transcriptional cell state, leading to a proximal enterocyte population enriched for genes within oxidative phosphorylation pathways. In addition, a population of active CD8+ T cells was recruited. Combined, this study provides a comprehensive profile of the p53 transcriptional response in vivo, revealing both tissue-specific transcriptomes and a unique signature, which were integrated to induce both cell-autonomous and non-cell-autonomous responses and transcriptional plasticity.
Keywords: Mdm2; p53; signature; single-cell sequencing; transcriptome.
Conflict of interest statement
The authors declare no competing interest.
Figures
References
-
- Vousden K. H., Prives C., Blinded by the light: The growing complexity of p53. Cell 137, 413–431 (2009). - PubMed
-
- Riley T., Sontag E., Chen P., Levine A., Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 9, 402–412 (2008). - PubMed
-
- Bensaad K. et al., TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell 126, 107–120 (2006). - PubMed
-
- Olivier M. et al., Recent advances in p53 research: An interdisciplinary perspective. Cancer Gene Ther. 16, 1–12 (2009). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

