Characterization of C- and D-Class MADS-Box Genes in Orchids
- PMID: 32900977
- PMCID: PMC7608164
- DOI: 10.1104/pp.20.00487
Characterization of C- and D-Class MADS-Box Genes in Orchids
Abstract
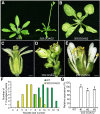
Orchids (members of the Orchidaceae family) possess unique flower morphology and adaptive reproduction strategies. Although the mechanisms underlying their perianth development have been intensively studied, the molecular basis of reproductive organ development in orchids remains largely unknown. Here, we report the identification and functional characterization of two AGAMOUS (AG)-like MADS-box genes, Dendrobium 'Orchid' AG1 (DOAG1) and DOAG2, which are putative C- and D-class genes, respectively, from the orchid Dendrobium 'Chao Praya Smile'. Both DOAG1 and DOAG2 are highly expressed in the reproductive organ, known as the column, compared to perianth organs, while DOAG2 expression gradually increases in pace with pollination-induced ovule development and is localized in ovule primordia. Ectopic expression of DOAG1, but not DOAG2, rescues floral defects in the Arabidopsis (Arabidopsis thaliana) ag-4 mutant, including reiteration of stamenoid perianth organs in inner whorls and complete loss of carpels. Downregulation of DOAG1 and DOAG2 in orchids by artificial microRNA interference using l-Met sulfoximine selection-based gene transformation systems shows that both genes are essential for specifying reproductive organ identity, yet they, exert different roles in mediating floral meristem determinacy and ovule development, respectively, in Dendrobium spp. orchids. Notably, knockdown of DOAG1 and DOAG2 also affects perianth organ development in orchids. Our findings suggest that DOAG1 and DOAG2 not only act as evolutionarily conserved C- and D-class genes, respectively, in determining reproductive organ identity, but also play hitherto unknown roles in mediating perianth organ development in orchids.
© 2020 American Society of Plant Biologists. All Rights Reserved.
Figures







References
-
- Carlson JE, Tulsieram LK, Glaubitz JC, Luk VWK, Kauffeldt C, Rutledge R(1991) Segregation of random amplified DNA markers in F1 progeny of conifers. Theor Appl Genet 83: 194–200 - PubMed
-
- Chai D, Lee SM, Ng JH, Yu H(2007) L-methionine sulfoximine as a novel selection agent for genetic transformation of orchids. J Biotechnol 131: 466–472 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

