Epitranscriptomics and epiproteomics in cancer drug resistance: therapeutic implications
- PMID: 32900991
- PMCID: PMC7479143
- DOI: 10.1038/s41392-020-00300-w
Epitranscriptomics and epiproteomics in cancer drug resistance: therapeutic implications
Abstract
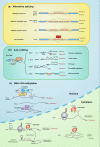
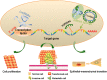
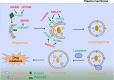
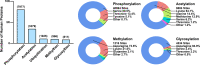
Drug resistance is a major hurdle in cancer treatment and a key cause of poor prognosis. Epitranscriptomics and epiproteomics are crucial in cell proliferation, migration, invasion, and epithelial-mesenchymal transition. In recent years, epitranscriptomic and epiproteomic modification has been investigated on their roles in overcoming drug resistance. In this review article, we summarized the recent progress in overcoming cancer drug resistance in three novel aspects: (i) mRNA modification, which includes alternative splicing, A-to-I modification and mRNA methylation; (ii) noncoding RNAs modification, which involves miRNAs, lncRNAs, and circRNAs; and (iii) posttranslational modification on molecules encompasses drug inactivation/efflux, drug target modifications, DNA damage repair, cell death resistance, EMT, and metastasis. In addition, we discussed the therapeutic implications of targeting some classical chemotherapeutic drugs such as cisplatin, 5-fluorouridine, and gefitinib via these modifications. Taken together, this review highlights the importance of epitranscriptomic and epiproteomic modification in cancer drug resistance and provides new insights on potential therapeutic targets to reverse cancer drug resistance.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Choi EK, et al. Body mass index and 20 specific cancers: re-analyses of dose-response meta-analyses of observational studies. Ann. Oncol. 2018;29:749–757. - PubMed
-
- Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. - PubMed
-
- Saitoh M. Involvement of partial EMT in cancer progression. J. Biochem. 2018;164:257–264. - PubMed
-
- Ayati A, et al. A review on progression of epidermal growth factor receptor (EGFR) inhibitors as an efficient approach in cancer targeted therapy. Bioorg. Chem. 2020;99:103811. - PubMed
-
- Sio TT, Ko J, Gudena VK, Verma N, Chaudhary UB. Chemotherapeutic and targeted biological agents for metastatic bladder cancer: a comprehensive review. Int J. Urol. 2014;21:630–637. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

