AiiDA 1.0, a scalable computational infrastructure for automated reproducible workflows and data provenance
- PMID: 32901044
- PMCID: PMC7479590
- DOI: 10.1038/s41597-020-00638-4
AiiDA 1.0, a scalable computational infrastructure for automated reproducible workflows and data provenance
Abstract
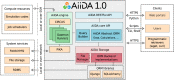
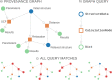
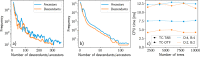
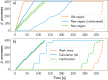
The ever-growing availability of computing power and the sustained development of advanced computational methods have contributed much to recent scientific progress. These developments present new challenges driven by the sheer amount of calculations and data to manage. Next-generation exascale supercomputers will harden these challenges, such that automated and scalable solutions become crucial. In recent years, we have been developing AiiDA (aiida.net), a robust open-source high-throughput infrastructure addressing the challenges arising from the needs of automated workflow management and data provenance recording. Here, we introduce developments and capabilities required to reach sustained performance, with AiiDA supporting throughputs of tens of thousands processes/hour, while automatically preserving and storing the full data provenance in a relational database making it queryable and traversable, thus enabling high-performance data analytics. AiiDA's workflow language provides advanced automation, error handling features and a flexible plugin model to allow interfacing with external simulation software. The associated plugin registry enables seamless sharing of extensions, empowering a vibrant user community dedicated to making simulations more robust, user-friendly and reproducible.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Stoddart, C. Is there a reproducibility crisis in science? Nat., 10.1038/d41586-019-00067-3 (2016).
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

