SRSF6 regulates alternative splicing of genes involved in DNA damage response and DNA repair in HeLa cells
- PMID: 32901876
- PMCID: PMC7551351
- DOI: 10.3892/or.2020.7750
SRSF6 regulates alternative splicing of genes involved in DNA damage response and DNA repair in HeLa cells
Abstract
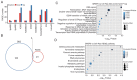
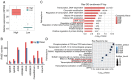
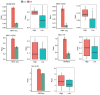
Alternative splicing (AS) occurs in nearly all human genes and abnormal AS has a close association with cancer. Serine and arginine‑rich splicing factor 6 (SRSF6), a canonical member of the serine/arginine‑rich protein family, has been characterized as an important regulator of AS. However, the role of SRSF6 in regulating AS in cancers has remained to be fully elucidated. In the present study, the median expression of SRSF6 in tumors was determined to be higher compared with that in matched normal tissues in 13 out of 16 cancer types from The Cancer Genome Atlas. To investigate the biological effects of SRSF6 overexpression, an SRSF6‑overexpression model of HeLa cells was constructed and it was revealed that SRSF6 overexpression resulted in significantly higher apoptosis and lower proliferation compared to control cells. Transcriptome analysis indicated that overexpression of SRSF6 in cancer cells induced large‑scale changes in transcriptional expression levels and AS. Two groups of cervical cancer tumor samples in which SRSF6 was differentially expressed were then selected to analyze potential SRSF6‑regulated AS. It was determined that the pattern of SRSF6‑regulated AS in clinical samples was similar to that in cancer cells and AS genes were enriched in DNA damage response (DDR) pathways, including DNA repair and double‑strand break repair via homologous recombination. Furthermore, AS events regulated by SRSF6 were validated using reverse transcription‑quantitative PCR. The present results highlighted that SRSF6 is able to trigger the activation of DDR pathways via regulation of AS to influence cancer progression. These results markedly expand the current understanding of the mechanisms underlying SRSF6‑mediated gene regulation and suggest the potential use of SRSF6 as a therapeutic target in cancer.
Keywords: SRSF6/SRP55; alternative splicing; cancer; DNA damage response; DNA repair.
Figures




References
-
- Kaczkowski B, Tanaka Y, Kawaji H, Sandelin A, Andersson R, Itoh M, Lassmann T, Hayashizaki Y, Carninci P, Forrest AR, FANTOM5 Consortium Transcriptome analysis of recurrently deregulated genes across multiple cancers identifies new pan-cancer biomarkers. Cancer Res. 2016;76:216–226. doi: 10.1158/0008-5472.CAN-15-0484. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

