On-Tissue Derivatization of Lipopolysaccharide for Detection of Lipid A Using MALDI-MSI
- PMID: 32902263
- PMCID: PMC8717242
- DOI: 10.1021/acs.analchem.0c02566
On-Tissue Derivatization of Lipopolysaccharide for Detection of Lipid A Using MALDI-MSI
Abstract
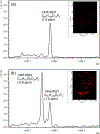
We developed a method to directly detect and map the Gram-negative bacterial virulence factor lipid A derived from lipopolysaccharide (LPS) by coupling acid hydrolysis with matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI-MSI). As the structure of lipid A (endotoxin) determines the innate immune outcome during infection, the ability to map its location within an infected organ or animal is needed to understand localized inflammatory responses that results during host-pathogen interactions. We previously demonstrated detection of free lipid A from infected tissue; however detection of lipid A derived from intact (smooth) LPS from host-pathogen MSI studies, proved elusive. Here, we detected LPS-derived lipid A from the Gram-negative pathogens, Escherichia coli (Ec, m/z 1797) and Pseudomonas aeruginosa (Pa, m/z 1446) using on-tissue acid hydrolysis to cleave the glycosidic linkage between the polysaccharide (core and O-antigen) and lipid A moieties of LPS. Using accurate mass methods, the ion corresponding to the major Ec and Pa lipid A species (m/z 1797 and 1446, respectively) were unambiguously discriminated from complex tissue substrates. Further, we evaluated potential delocalization and signal loss of other tissue lipids and found no evidence for either, making this LPS-to-Lipid A-MSI (LLA-MSI) method, compatible with simultaneous host-pathogen lipid imaging following acid hydrolysis. This spatially sensitive technique is the first step in mapping host-influenced de novo lipid A modifications, such as those associated with antimicrobial resistance phenotypes, during Gram-negative bacterial infection and will advance our understanding of the host-pathogen interface.
Figures

References
-
- Castellino S; Groseclose MR; Wagner D Bioanalysis 2011, 3 (21), 2427–2441. - PubMed
-
- Schulz S; Becker M; Groseclose MR; Schadt S; Hopf C Curr. Opin. Biotechnol 2019, 55, 51–59. - PubMed
-
- Amstalden van Hove ER; Smith DF; Heeren RMA Journal of Chromatography A 2010, 1217 (25), 3946–3954. - PubMed
-
- Cornett DS; Reyzer ML; Chaurand P; Caprioli RM Nat. Methods 2007, 4 (10), 828–833. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

