Peracylation Coupled with Tandem Mass Spectrometry for Structural Sequencing of Sulfated Glycosaminoglycan Mixtures without Depolymerization
- PMID: 32902282
- PMCID: PMC7664153
- DOI: 10.1021/jasms.0c00178
Peracylation Coupled with Tandem Mass Spectrometry for Structural Sequencing of Sulfated Glycosaminoglycan Mixtures without Depolymerization
Abstract
The structures of glycosaminoglycans (GAGs), especially the patterns of modification, are crucial to modulate interactions with various protein targets. It is very challenging to determine the fine structures using liquid chromatography-mass spectrometry (LC-MS) due in large part to the gas-phase sulfate losses upon collisional activation. Previously, our group reported a method for fine structure analysis that required permethylation of the GAG oligosaccharide. However, uncontrolled depolymerization during the permethylation process due to esterification of uronic acid lowers the reliability of the method to resolve structures of GAGs, especially for larger oligosaccharides. Here, we describe a simplified derivatization method using propionylation and desulfation. The oligosaccharides have all hydroxyl and amine groups protected with propionyl groups and then have sulfate groups removed to generate unprotected hydroxyl and amine groups at all sites that were previously sulfated. This derivatized oligosaccharide generates informative fragments during collision-induced dissociation that resolve the original sulfation patterns. This method is demonstrated to enable accurate determination of sulfation patterns of even the highly sulfated pentasaccharide fondaparinux by MS2 and MS3. Using a mixture of dp6 from porcine heparin, we demonstrate that this method allows for structural characterization of complex mixtures, including clear chromatographic separation and sequencing of structural isomers, all at high yields without evidence of depolymerization. This represents a marked improvement in the reliability to structurally characterize GAG oligosaccharides over permethylation-based derivatization schemes.
Keywords: LC-MS/MS; carbohydrates; derivatization; glycosaminoglycans; heparin.
Figures
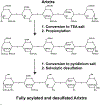


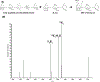
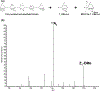
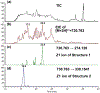
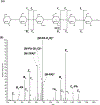
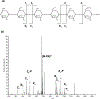
References
-
- Taylor KR, Gallo RL. Glycosaminoglycans and their proteoglycans: host-associated molecular patterns for initiation and modulation of inflammation. FASEB J 20(1), 9–22 (2006) - PubMed
-
- Gandhi NS, Mancera RL. The structure of glycosaminoglycans and their interactions with proteins. Chem Biol Drug Des 72(6), 455–482 (2008) - PubMed
-
- Capila I, Linhardt RJ. Heparin–protein interactions. Angew. Chem. Int. Ed. Engl 41(3), 390–412 (2002) - PubMed
-
- Rabenstein DL. Heparin and heparan sulfate: structure and function. Nat. Prod. Rep 19(3), 312–331 (2002) - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

