mRNA and miRNA profiling of Zika virus-infected human umbilical cord mesenchymal stem cells identifies miR-142-5p as an antiviral factor
- PMID: 32902370
- PMCID: PMC7534337
- DOI: 10.1080/22221751.2020.1821581
mRNA and miRNA profiling of Zika virus-infected human umbilical cord mesenchymal stem cells identifies miR-142-5p as an antiviral factor
Abstract
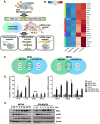
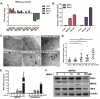
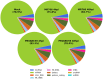
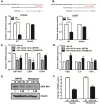
Zika virus (ZIKV) infection during pregnancy is associated with congenital brain abnormalities, a finding that highlights the urgent need to understand mother-to-fetus transmission mechanisms. Human umbilical cord mesenchymal stem cells (hUCMSCs) are susceptible to ZIKV infection but the underlying mechanisms of viral susceptibility remain largely unexplored. In this study, we have characterized and compared host mRNA and miRNA expression profiles in hUCMSCs after infection with two lineages of ZIKV, African (MR766) and Asian (PRVABC59). RNA sequencing analysis identified differentially expressed genes involved in anti-viral immunity and mitochondrial dynamics following ZIKV infection. In particular, ZIKV-infected hUCMSCs displayed mitochondrial elongation and the treatment of hUCMSCs with mitochondrial fission inhibitor led to a dose-dependent increase in ZIKV gene expression and decrease in anti-viral signalling pathways. Moreover, small RNA sequencing analysis identified several significantly up- or down-regulated microRNAs. Interestingly, miR-142-5p was significantly downregulated upon ZIKV infection, whereas cellular targets of miR-142-5p, IL6ST and ITGAV, were upregulated. Overexpression of miR-142-5p resulted in the suppression of ZIKV replication. Furthermore, blocking ITGAV expression resulted in a significant suppression of ZIKV binding to cells, suggesting a potential role of ITGAV in ZIKV entry. In conclusion, these results demonstrate both common and specific host responses to African and Asian ZIKV lineages and indicate miR-142-5p as a key regulator of ZIKV replication in the umbilical cords.
Keywords: RNA-seq; ZIKV; hUCMSCs; innate immunity; miRNA; small RNA-seq.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



Similar articles
-
Integrated MicroRNA and mRNA Profiling in Zika Virus-Infected Neurons.Viruses. 2019 Feb 16;11(2):162. doi: 10.3390/v11020162. Viruses. 2019. PMID: 30781519 Free PMC article.
-
Robust antiviral innate immune response and miRNA regulatory network were identified in ZIKV-infected cells: implications in the pathogenesis of ZIKV infection.Virus Genes. 2025 Jun;61(3):249-264. doi: 10.1007/s11262-025-02136-4. Epub 2025 Feb 16. Virus Genes. 2025. PMID: 39955676
-
Insights into ZIKV-Mediated Innate Immune Responses in Human Dermal Fibroblasts and Epidermal Keratinocytes.J Invest Dermatol. 2019 Feb;139(2):391-399. doi: 10.1016/j.jid.2018.07.038. Epub 2018 Sep 12. J Invest Dermatol. 2019. PMID: 30218650
-
MicroRNA profiles in Zika virus infection: Insights from diverse sources.Indian J Med Microbiol. 2024 Sep-Oct;51:100697. doi: 10.1016/j.ijmmb.2024.100697. Epub 2024 Aug 5. Indian J Med Microbiol. 2024. PMID: 39103054 Review.
-
Evasion of Innate and Intrinsic Antiviral Pathways by the Zika Virus.Viruses. 2019 Oct 22;11(10):970. doi: 10.3390/v11100970. Viruses. 2019. PMID: 31652496 Free PMC article. Review.
Cited by
-
RNA-Seq analysis of duck embryo fibroblast cells gene expression during duck Tembusu virus infection.Vet Res. 2022 May 18;53(1):34. doi: 10.1186/s13567-022-01051-y. Vet Res. 2022. PMID: 35585616 Free PMC article.
-
Regulatory Role of Host MicroRNAs in Flaviviruses Infection.Front Microbiol. 2022 Apr 11;13:869441. doi: 10.3389/fmicb.2022.869441. eCollection 2022. Front Microbiol. 2022. PMID: 35479613 Free PMC article. Review.
-
MiR-103a-3p Promotes Zika Virus Replication by Targeting OTU Deubiquitinase 4 to Activate p38 Mitogen-Activated Protein Kinase Signaling Pathway.Front Microbiol. 2022 Mar 4;13:862580. doi: 10.3389/fmicb.2022.862580. eCollection 2022. Front Microbiol. 2022. PMID: 35317262 Free PMC article.
-
Nonstructural Protein NS1 of Influenza Virus Disrupts Mitochondrial Dynamics and Enhances Mitophagy via ULK1 and BNIP3.Viruses. 2021 Sep 15;13(9):1845. doi: 10.3390/v13091845. Viruses. 2021. PMID: 34578425 Free PMC article.
-
SARS-CoV-2 Nucleocapsid Protein Targets RIG-I-Like Receptor Pathways to Inhibit the Induction of Interferon Response.Cells. 2021 Mar 2;10(3):530. doi: 10.3390/cells10030530. Cells. 2021. PMID: 33801464 Free PMC article.
References
-
- Dick GW, Kitchen SF, Haddow AJ.. Zika virus. I. Isolations and serological specificity. Trans R Soc Trop Med Hyg. 1952 Sep;46(5):509–520. - PubMed
-
- Dyer O. Zika virus spreads across Americas as concerns mount over birth defects. Br Med J. 2015 Dec 23;351:h6983. - PubMed
-
- Fauci AS, Morens DM.. Zika virus in the Americas – yet another Arbovirus threat. N Engl J Med. 2016 Feb 18;374(7):601–604. - PubMed
-
- Mlakar J, Korva M, Tul N, et al. . Zika virus associated with microcephaly. N Engl J Med. 2016 Mar 10;374(10):951–958. - PubMed
-
- McCarthy M. Zika related microcephaly may appear after birth, study finds. Br Med J. 2016 Nov 23;355:i6333. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
