Predicting the recombination potential of severe acute respiratory syndrome coronavirus 2 and Middle East respiratory syndrome coronavirus
- PMID: 32902372
- PMCID: PMC7819352
- DOI: 10.1099/jgv.0.001491
Predicting the recombination potential of severe acute respiratory syndrome coronavirus 2 and Middle East respiratory syndrome coronavirus
Abstract
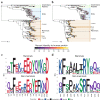
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) recently emerged to cause widespread infections in humans. SARS-CoV-2 infections have been reported in the Kingdom of Saudi Arabia, where Middle East respiratory syndrome coronavirus (MERS-CoV) causes seasonal outbreaks with a case fatality rate of ~37 %. Here we show that there exists a theoretical possibility of future recombination events between SARS-CoV-2 and MERS-CoV RNA. Through computational analyses, we have identified homologous genomic regions within the ORF1ab and S genes that could facilitate recombination, and have analysed co-expression patterns of the cellular receptors for SARS-CoV-2 and MERS-CoV, ACE2 and DPP4, respectively, to identify human anatomical sites that could facilitate co-infection. Furthermore, we have investigated the likely susceptibility of various animal species to MERS-CoV and SARS-CoV-2 infection by comparing known virus spike protein-receptor interacting residues. In conclusion, we suggest that a recombination between SARS-CoV-2 and MERS-CoV RNA is possible and urge public health laboratories in high-risk areas to develop diagnostic capability for the detection of recombined coronaviruses in patient samples.
Keywords: MERS-CoV; SARS-CoV-2; coronavirus; emergence; predictions; recombination.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

