Transforming growth factor beta signaling and decidual integrity in mice†
- PMID: 32902612
- PMCID: PMC7711917
- DOI: 10.1093/biolre/ioaa155
Transforming growth factor beta signaling and decidual integrity in mice†
Abstract
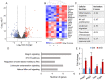
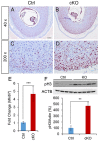
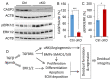
Transforming growth factor beta (TGFβ) signaling regulates multifaceted reproductive processes. It has been shown that the type 1 receptor of TGFβ (TGFBR1) is indispensable for female reproductive tract development, implantation, placental development, and fertility. However, the role of TGFβ signaling in decidual development and function remains poorly defined. Our objective is to determine the impact of uterine-specific deletion of Tgfbr1 on decidual integrity, with a focus on the cellular and molecular properties of the decidua during development. Our results show that the developmental dynamics of the decidua is altered in TGFBR1 conditionally depleted uteri from embryonic day (E) 5.5 to E8.5, substantiated by downregulation of genes associated with inflammatory responses and uterine natural killer cell abundance, reduced presence of nondecidualized fibroblasts in the antimesometrial region, and altered decidual cell development. Notably, conditional ablation of TGFBR1 results in the formation of decidua containing more abundant alpha smooth muscle actin (ACTA2)-positive cells at the peripheral region of the antimesometrial side versus controls at E6.5-E8.5. This finding is corroborated by upregulation of a subset of smooth muscle marker genes in Tgfbr1 conditionally deleted decidua at E6.5 and E8.5. Moreover, increased cell proliferation and enhanced decidual ERK1/2 signaling were found in Tgfbr1 conditional knockout mice upon decidual regression. In summary, conditional ablation of TGFBR1 in the uterus profoundly impacts the cellular and molecular properties of the decidua. Our results suggest that TGFBR1 in uterine epithelial and stromal compartments is important for the integrity of the decidua, a transient but crucial structure that supports embryo development.
Keywords: TGFBR1; TGFβ signaling; decidualization; endometrium; uterus.
© The Author(s) 2020. Published by Oxford University Press on behalf of Society for the Study of Reproduction. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

