Targeting a cytokine checkpoint enhances the fitness of armored cord blood CAR-NK cells
- PMID: 32902645
- PMCID: PMC7869185
- DOI: 10.1182/blood.2020007748
Targeting a cytokine checkpoint enhances the fitness of armored cord blood CAR-NK cells
Erratum in
-
Daher M, Basar R, Gokdemir E, et al. Targeting a cytokine checkpoint enhances the fitness of armored cord blood CAR-NK cells. Blood. 2021;137(5):624-636.Blood. 2023 Mar 9;141(10):1235. doi: 10.1182/blood.2023019760. Blood. 2023. PMID: 36893002 Free PMC article. No abstract available.
Abstract
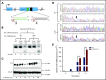
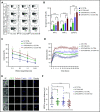
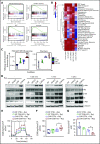
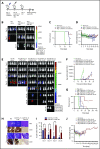
Immune checkpoint therapy has resulted in remarkable improvements in the outcome for certain cancers. To broaden the clinical impact of checkpoint targeting, we devised a strategy that couples targeting of the cytokine-inducible Src homology 2-containing (CIS) protein, a key negative regulator of interleukin 15 (IL-15) signaling, with fourth-generation "armored" chimeric antigen receptor (CAR) engineering of cord blood-derived natural killer (NK) cells. This combined strategy boosted NK cell effector function through enhancing the Akt/mTORC1 axis and c-MYC signaling, resulting in increased aerobic glycolysis. When tested in a lymphoma mouse model, this combined approach improved NK cell antitumor activity more than either alteration alone, eradicating lymphoma xenografts without signs of any measurable toxicity. We conclude that targeting a cytokine checkpoint further enhances the antitumor activity of IL-15-secreting armored CAR-NK cells by promoting their metabolic fitness and antitumor activity. This combined approach represents a promising milestone in the development of the next generation of NK cells for cancer immunotherapy.
© 2021 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: K.R., M.D., R.B., P.P.B., E.L., E.J.S, R.E.C., D.M., and The University of Texas MD Anderson Cancer Center (MDACC) have an institutional financial conflict of interest with Takeda Pharmaceutical for the licensing of the technology related to the research reported here. MDACC has implemented an Institutional Conflict of Interest Management and Monitoring Plan to manage and monitor the conflict of interest with respect to MDACC’s conduct of any other ongoing or future research related to this relationship. M.A.B., M.S.S., G.R.R., M.S.M., G.K., and R.T. are employed by IDT, which manufactures reagents similar to some described in the paper; M.A.B., M.S.M., R.T., and G.R.R. own equity in DHR, the parent company of IDT. H.L. is a consultant for IDT. The remaining authors declare no competing financial interests.
Figures



Comment in
-
Releasing the brake in CAR natural killer cells.Blood. 2021 Feb 4;137(5):579-581. doi: 10.1182/blood.2020008993. Blood. 2021. PMID: 33538803 No abstract available.
References
-
- Morvan MG, Lanier LL. NK cells and cancer: you can teach innate cells new tricks. Nat Rev Cancer. 2016;16(1):7-19. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

