Antivirals Against Coronaviruses: Candidate Drugs for SARS-CoV-2 Treatment?
- PMID: 32903349
- PMCID: PMC7438404
- DOI: 10.3389/fmicb.2020.01818
Antivirals Against Coronaviruses: Candidate Drugs for SARS-CoV-2 Treatment?
Abstract
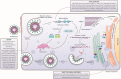
Coronaviruses (CoVs) are a group of viruses from the family Coronaviridae that can infect humans and animals, causing mild to severe diseases. The ongoing pandemic of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) represents a global threat, urging the development of new therapeutic strategies. Here we present a selection of relevant compounds that have been described from 2005 until now as having in vitro and/or in vivo antiviral activities against human and/or animal CoVs. We also present compounds that have reached clinical trials as well as further discussing the potentiality of other molecules for application in (re)emergent CoVs outbreaks. Finally, through rationalization of the data presented herein, we wish to encourage further research encompassing these compounds as potential SARS-CoV-2 drug candidates.
Keywords: COVID-19; SARS-CoV-2; antivirals; coronaviruses; treatment.
Copyright © 2020 Santos, Grosche, Bergamini, Sabino-Silva and Jardim.
Figures

References
-
- Agnihothram S., Gopal R., Yount B. L., Donaldson E. F., Menachery V. D., Graham R. L., et al. (2014). Evaluation of serologic and antigenic relationships between middle eastern respiratory syndrome coronavirus and other coronaviruses to develop vaccine platforms for the rapid response to emerging coronaviruses. J. Infect. Dis. 209 995–1006. 10.1093/infdis/jit609 - DOI - PMC - PubMed
-
- Aguiar A. C. C., Murce E., Cortopassi W. A., Pimentel A. S., Almeida M. M. F. S., Barros D. C. S., et al. (2018). Chloroquine analogs as antimalarial candidates with potent in vitro and in vivo activity. Int. J. Parasitol. Drugs Drug Resist. 8 459–464. 10.1016/j.ijpddr.2018.10.002 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

