Transcriptomic Analysis of Rhodococcus opacus R7 Grown on o-Xylene by RNA-Seq
- PMID: 32903390
- PMCID: PMC7434839
- DOI: 10.3389/fmicb.2020.01808
Transcriptomic Analysis of Rhodococcus opacus R7 Grown on o-Xylene by RNA-Seq
Abstract
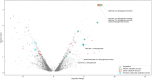
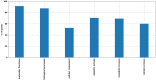
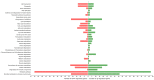
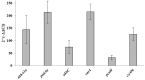
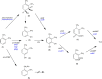
Xylenes are considered one of the most common hazardous sources of environmental contamination. The biodegradation of these compounds has been often reported, rarer the ability to oxidize the ortho-isomer. Among few o-xylene-degrading bacteria, Rhodococcus opacus R7 is well known for its capability to degrade diverse aromatic hydrocarbons and toxic compounds, including o-xylene as only carbon and energy source. This work shows for the first time the RNA-seq approach to elucidate the genetic determinants involved in the o-xylene degradation pathway in R. opacus R7. Transcriptomic data showed 542 differentially expressed genes that are associated with the oxidation of aromatic hydrocarbons and stress response, osmotic regulation and central metabolism. Gene ontology (GO) enrichment and KEGG pathway analysis confirmed significant changes in aromatic compound catabolic processes, fatty acid metabolism, beta-oxidation, TCA cycle enzymes, and biosynthesis of metabolites when cells are cultured in the presence of o-xylene. Interestingly, the most up-regulated genes belong to the akb gene cluster encoding for the ethylbenzene (Akb) dioxygenase system. Moreover, the transcriptomic approach allowed identifying candidate enzymes involved in R7 o-xylene degradation for their likely participation in the formation of the metabolites that have been previously identified. Overall, this approach supports the identification of several oxidative systems likely involved in o-xylene metabolism confirming that R. opacus R7 possesses a redundancy of sequences that converge in o-xylene degradation through R7 peculiar degradation pathway. This work advances our understanding of o-xylene metabolism in bacteria belonging to Rhodococcus genus and provides a framework of useful enzymes (molecular tools) that can be fruitfully targeted for optimized o-xylene consumption.
Keywords: RNA-seq; Rhodococcus opacus; environmental contamination; o-xylene; oxygenases; stress response.
Copyright © 2020 Zampolli, Di Canito, Manconi, Milanesi, Di Gennaro and Orro.
Figures
References
-
- Alvarez H. M. (2019). Biology of Rhodococcus. Berlin: Springer; 10.1007/978-3-030-11461-9 - DOI
-
- Barbieri P., Palladino L., Di Gennaro P., Galli E. (1993). Alternative pathways for o-xylene or m-xylene and p-xylene degradation in Pseudomonas stutzeri strain. Biodegradation 4 71–80. 10.1007/BF00702323 - DOI

