Hsa-miR-155-5p Up-Regulation in Breast Cancer and Its Relevance for Treatment With Poly[ADP-Ribose] Polymerase 1 (PARP-1) Inhibitors
- PMID: 32903519
- PMCID: PMC7435065
- DOI: 10.3389/fonc.2020.01415
Hsa-miR-155-5p Up-Regulation in Breast Cancer and Its Relevance for Treatment With Poly[ADP-Ribose] Polymerase 1 (PARP-1) Inhibitors
Abstract
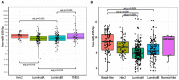
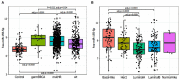
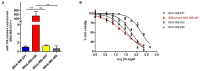
miR-155-5p is a well-known oncogenic microRNA, showing frequent overexpression in human malignancies, including breast cancer. Here, we show that high miR-155-5p levels are associated with unfavorable prognostic factors in two independent breast cancer cohorts (CSS cohort, n = 283; and TCGA-BRCA dataset, n = 1,095). Consistently, miR-155-5p results as differentially expressed in the breast cancer subgroups identified by the surrogate molecular classification in the CSS cohort and the PAM50 classifier in TCGA-BRCA dataset, with the TNBC and HER2-amplified tumors carrying the highest levels. Since the analysis of TCGA-BC dataset also demonstrated a significant association between miR-155-5p levels and the presence of mutations in homologous recombination (HR) genes, we hypothesized that miR-155-5p might affect cell response to the PARP-1 inhibitor Olaparib. As expected, miR-155-5p ectopic overexpression followed by Olaparib administration resulted in a greater reduction of cell viability as compared to Olaparib administration alone, suggesting that miR-155-5p might induce a synthetic lethal effect in cancer cells when coupled with PARP-1-inhibition. Overall, our data point to a role of miR-155-5p in homologous recombination deficiency and suggest miR-155-5p might be useful in predicting response to PARP1 inhibitors in the clinical setting.
Keywords: Olaparib; PARP-1 inhibitors; breast cancer; homologous recombination; hsa-miR-155-5p.
Copyright © 2020 Pasculli, Barbano, Fontana, Biagini, Di Viesti, Rendina, Valori, Morritti, Bravaccini, Ravaioli, Maiello, Graziano, Murgo, Copetti, Mazza, Fazio, Esteller and Parrella.
Figures


References
-
- Curigliano G, Burstein HJ, P Winer E, Gnant M, Dubsky P, Loibl S, et al. . De-escalating and escalating treatments for early-stage breast cancer: the St. Gallen International Expert Consensus Conference on the Primary Therapy of Early Breast Cancer 2017. Ann Oncol. (2019) 30:1181. 10.1093/annonc/mdy537 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

