RNA-Seq-Based Whole Transcriptome Analysis of IPEC-J2 Cells During Swine Acute Diarrhea Syndrome Coronavirus Infection
- PMID: 32903570
- PMCID: PMC7438718
- DOI: 10.3389/fvets.2020.00492
RNA-Seq-Based Whole Transcriptome Analysis of IPEC-J2 Cells During Swine Acute Diarrhea Syndrome Coronavirus Infection
Abstract
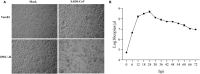
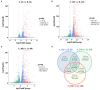
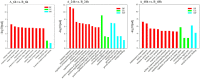
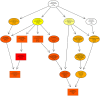
The new emergence of swine acute diarrhea syndrome coronavirus (SADS-CoV) has resulted in high mortality in suckling pigs in China. To date, the transcriptional expression of host cells during SADS-CoV infection has not been documented. In this study, by means of RNA-Seq technology, we investigated the whole genomic expression profiles of intestinal porcine epithelial cells (IPEC-J2) infected with a SADS-CoV strain SADS-CoV-CH-FJWT-2018. A total of 24,676 genes were identified: 23,677 were known genes, and 999 were novel genes. A total of 1,897 differentially expressed genes (DEGs) were identified between SADS-CoV-infected and uninfected cells at 6, 24, and 48 h post infection (hpi). Of these, 1,260 genes were upregulated and 637 downregulated. A Gene Ontology enrichment analysis revealed that DEGs in samples from 6, 24, and 48 hpi were enriched in 79, 383, and 233 GO terms, respectively, which were mainly involved in immune system process, response to stimulus, signal transduction, and cytokine-cytokine receptor interactions. The 1,897 DEGs were mapped to 109 KEGG Ontology (KO) pathways classified into four main categories. Most of the DEGs annotated in the KEGG pathways were related to the immune system, infectious viral disease, and signal transduction. The mRNA of porcine serum amyloid A-3 protein (SAA3), an acute phase response protein, was significantly upregulated during the infection. Over-expressed SAA3 in IPEC-J2 cells drastically inhibited the replication of SADS-CoV, while under-expressed SAA3 promoted virus replication. To our knowledge, this is the first report on the profiles of gene expression of IPEC-J2 cells infected by SADS-CoV by means of RNA-Seq technology. Our results indicate that SADS-CoV infection significantly modified the host cell gene expression patterns, and the host cells responded in highly specific manners, including immune response, signal and cytokine transduction, and antiviral response. The findings provide important insights into the transcriptome of IPEC-J2 in SADS-CoV infection.
Keywords: RNA-Seq; SADS-CoV; gene expression; porcine serum amyloid A-3; transcriptome.
Copyright © 2020 Zhang, Yuan, Li, Zhang, Ye, Li, Ding, Chen, Cheng, Wu, Tang and Song.
Figures


Similar articles
-
Transcriptional Landscape of Vero E6 Cells during Early Swine Acute Diarrhea Syndrome Coronavirus Infection.Viruses. 2021 Apr 14;13(4):674. doi: 10.3390/v13040674. Viruses. 2021. PMID: 33919952 Free PMC article.
-
Comparative transcriptomic analysis of porcine epidemic diarrhea virus epidemic and classical strains in IPEC-J2 cells.Vet Microbiol. 2022 Oct;273:109540. doi: 10.1016/j.vetmic.2022.109540. Epub 2022 Aug 8. Vet Microbiol. 2022. PMID: 35987184
-
Transcriptome analysis of host response to porcine epidemic diarrhea virus nsp15 in IPEC-J2 cells.Microb Pathog. 2022 Jan;162:105195. doi: 10.1016/j.micpath.2021.105195. Epub 2021 Sep 24. Microb Pathog. 2022. PMID: 34571150
-
Swine Acute Diarrhea Syndrome Coronavirus: An Overview of Virus Structure and Virus-Host Interactions.Animals (Basel). 2025 Jan 9;15(2):149. doi: 10.3390/ani15020149. Animals (Basel). 2025. PMID: 39858149 Free PMC article. Review.
-
Research Advances on Swine Acute Diarrhea Syndrome Coronavirus.Animals (Basel). 2024 Jan 30;14(3):448. doi: 10.3390/ani14030448. Animals (Basel). 2024. PMID: 38338091 Free PMC article. Review.
Cited by
-
LACpG10-HL Functions Effectively in Antibiotic-Free and Healthy Husbandry by Improving the Innate Immunity.Int J Mol Sci. 2022 Sep 28;23(19):11466. doi: 10.3390/ijms231911466. Int J Mol Sci. 2022. PMID: 36232768 Free PMC article.
-
Transcriptional Landscape of Vero E6 Cells during Early Swine Acute Diarrhea Syndrome Coronavirus Infection.Viruses. 2021 Apr 14;13(4):674. doi: 10.3390/v13040674. Viruses. 2021. PMID: 33919952 Free PMC article.
-
Differing coronavirus genres alter shared host signaling pathways upon viral infection.Sci Rep. 2022 Jun 13;12(1):9744. doi: 10.1038/s41598-022-13396-7. Sci Rep. 2022. PMID: 35697915 Free PMC article.
-
Describing variability in pig genes involved in coronavirus infections for a One Health perspective in conservation of animal genetic resources.Sci Rep. 2021 Feb 9;11(1):3359. doi: 10.1038/s41598-021-82956-0. Sci Rep. 2021. PMID: 33564056 Free PMC article.
-
Remdesivir inhibits Porcine epidemic diarrhea virus infection in vitro.Heliyon. 2023 Nov 2;9(11):e21468. doi: 10.1016/j.heliyon.2023.e21468. eCollection 2023 Nov. Heliyon. 2023. PMID: 38027806 Free PMC article.
References
-
- Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, Lau JH, et al. . Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. (2012) 86:3995–4008. 10.1128/JVI.06540-11 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

