Unbiased Characterization of Peptide-HLA Class II Interactions Based on Large-Scale Peptide Microarrays; Assessment of the Impact on HLA Class II Ligand and Epitope Prediction
- PMID: 32903714
- PMCID: PMC7438773
- DOI: 10.3389/fimmu.2020.01705
Unbiased Characterization of Peptide-HLA Class II Interactions Based on Large-Scale Peptide Microarrays; Assessment of the Impact on HLA Class II Ligand and Epitope Prediction
Abstract
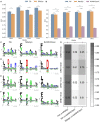
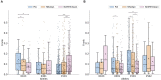
Human Leukocyte Antigen class II (HLA-II) molecules present peptides to T lymphocytes and play an important role in adaptive immune responses. Characterizing the binding specificity of single HLA-II molecules has profound impacts for understanding cellular immunity, identifying the cause of autoimmune diseases, for immunotherapeutics, and vaccine development. Here, novel high-density peptide microarray technology combined with machine learning techniques were used to address this task at an unprecedented level of high-throughput. Microarrays with over 200,000 defined peptides were assayed with four exemplary HLA-II molecules. Machine learning was applied to mine the signals. The comparison of identified binding motifs, and power for predicting eluted ligands and CD4+ epitope datasets to that obtained using NetMHCIIpan-3.2, confirmed a high quality of the chip readout. These results suggest that the proposed microarray technology offers a novel and unique platform for large-scale unbiased interrogation of peptide binding preferences of HLA-II molecules.
Keywords: HLA; MHC class II; antigen presentation; high-throughput; machine learning; peptide binding; prediction; ultra-high density peptide microarray.
Copyright © 2020 Wendorff, Garcia Alvarez, Østerbye, ElAbd, Rosati, Degenhardt, Buus, Franke and Nielsen.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

