The Positive Lymph Node Ratio Predicts Survival in T1-4N1-3M0 Non-Small Cell Lung Cancer: A Nomogram Using the SEER Database
- PMID: 32903785
- PMCID: PMC7438846
- DOI: 10.3389/fonc.2020.01356
The Positive Lymph Node Ratio Predicts Survival in T1-4N1-3M0 Non-Small Cell Lung Cancer: A Nomogram Using the SEER Database
Abstract
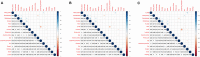
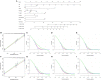
Background: An increasing number of studies have shown that the positive lymph node ratio (pLNR) can be used to evaluate the prognosis of non-small cell lung cancer (NSCLC) patients. To determine the predictive value of the pLNR, we collected data from the Surveillance, Epidemiology, and End Results (SEER) database and performed a retrospective analysis. Methods: We collected survival and clinical information on patients with T1-4N1-3M0 NSCLC diagnosed between 2010 and 2016 from the SEER database and screened them according to inclusion and exclusion criteria. X-tile software was used to obtain the best cut-off value for the pLNR. Then, we randomly divided patients into a training set and a validation set at a ratio of 7:3. Pearson's correlation coefficient, tolerance and the variance inflation factor (VIF) were used to detect collinearity between variables. Univariate and multivariate Cox regression analyses were used to identify significant prognostic factors, and nomograms was constructed to visualize the results. The concordance index (C-index), calibration curves, and decision curve analysis (DCA) were used to assess the predictive ability of the nomogram. We divided the patient scores into four groups according to the interquartile interval and constructed a survival curve using Kaplan-Meier analysis. Results: A total of 6,245 patients were initially enrolled. The best cut-off value for the pLNR was determined to be 0.55. The nomogram contained 13 prognostic factors, including the pLNR. The pLNR was identified as an independent prognostic factor for both overall survival (OS) and cancer-specific survival (CSS). The C-index was 0.703 (95% CI, 0.695-0.711) in the training set and 0.711 (95% CI, 0.699-0.723) in the validation set. The calibration curves and DCA also indicated the good predictability of the nomogram. Risk stratification revealed a statistically significant difference among the four groups of patients divided according to quartiles of risk score. Conclusion: The nomogram containing the pLNR can accurately predict survival in patients with T1-4N1-3M0 NSCLC.
Keywords: SEER; nomogram; non-small cell lung cancer; positive lymph node; prognosis.
Copyright © 2020 Liao, Yin and Fan.
Figures





References
-
- Rami-Porta R, Bolejack V, Crowley J, Ball D, Kim J, Lyons G, et al. The IASLC lung cancer staging project: proposals for the revisions of the T descriptors in the forthcoming eighth edition of the TNM classification for lung cancer. J Thorac Oncol. (2015) 10:990–1003. 10.1097/JTO.0000000000000559 - DOI - PubMed
LinkOut - more resources
Full Text Sources

