Comparative Genomics of Microbacterium Species to Reveal Diversity, Potential for Secondary Metabolites and Heavy Metal Resistance
- PMID: 32903828
- PMCID: PMC7438953
- DOI: 10.3389/fmicb.2020.01869
Comparative Genomics of Microbacterium Species to Reveal Diversity, Potential for Secondary Metabolites and Heavy Metal Resistance
Abstract
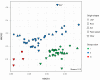
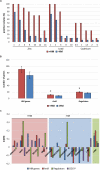
Microbacterium species have been isolated from a wide range of hosts and environments, including heavy metal-contaminated sites. Here, we present a comprehensive analysis on the phylogenetic distribution and the genetic potential of 70 Microbacterium belonging to 20 different species isolated from heavy metal-contaminated and non-contaminated sites with particular attention to secondary metabolites gene clusters. The analyzed Microbacterium species are divided in three main functional clades. They share a small core genome (331 gene families covering basic functions) pointing to high genetic diversity. The most common secondary metabolite gene clusters encode pathways for the production of terpenoids, type III polyketide synthases and non-ribosomal peptide synthetases, potentially responsible of the synthesis of siderophore-like compounds. In vitro tests showed that many Microbacterium strains produce siderophores, ACC deaminase, auxins (IAA) and are able to solubilize phosphate. Microbacterium isolates from heavy metal contaminated sites are on average more resistant to heavy metals and harbor more genes related to metal homeostasis (e.g., metalloregulators). On the other hand, the ability to increase the metal mobility in a contaminated soil through the secretion of specific molecules seems to be widespread among all. Despite the widespread capacity of strains to mobilize several metals, plants inoculated with selected Microbacterium isolates showed only slightly increased iron concentrations, whereas concentrations of zinc, cadmium and lead were decreased.
Keywords: comparative genomics; heavy metals; plant associated bacteria; polyketide synthases; secondary metabolites; siderophore; terpenoids.
Copyright © 2020 Corretto, Antonielli, Sessitsch, Höfer, Puschenreiter, Widhalm, Swarnalakshmi and Brader.
Figures


Similar articles
-
Comparative genomics of 16 Microbacterium spp. that tolerate multiple heavy metals and antibiotics.PeerJ. 2019 Jan 14;6:e6258. doi: 10.7717/peerj.6258. eCollection 2019. PeerJ. 2019. PMID: 30671291 Free PMC article.
-
The hyperaccumulator Sedum plumbizincicola harbors metal-resistant endophytic bacteria that improve its phytoextraction capacity in multi-metal contaminated soil.J Environ Manage. 2015 Jun 1;156:62-9. doi: 10.1016/j.jenvman.2015.03.024. Epub 2015 Mar 19. J Environ Manage. 2015. PMID: 25796039
-
Arsenic Hyper-tolerance in Four Microbacterium Species Isolated from Soil Contaminated with Textile Effluent.Toxicol Int. 2012 May;19(2):188-94. doi: 10.4103/0971-6580.97221. Toxicol Int. 2012. PMID: 22778519 Free PMC article.
-
Potential of siderophore-producing bacteria for improving heavy metal phytoextraction.Trends Biotechnol. 2010 Mar;28(3):142-9. doi: 10.1016/j.tibtech.2009.12.002. Epub 2010 Jan 13. Trends Biotechnol. 2010. PMID: 20044160 Review.
-
Advances in the application of plant growth-promoting rhizobacteria in phytoremediation of heavy metals.Rev Environ Contam Toxicol. 2013;223:33-52. doi: 10.1007/978-1-4614-5577-6_2. Rev Environ Contam Toxicol. 2013. PMID: 23149811 Review.
Cited by
-
Description of Microbacterium luteum sp. nov., Microbacterium cremeum sp. nov., and Microbacterium atlanticum sp. nov., three novel C50 carotenoid producing bacteria.J Microbiol. 2021 Oct;59(10):886-897. doi: 10.1007/s12275-021-1186-5. Epub 2021 Sep 7. J Microbiol. 2021. PMID: 34491524
-
Biodesulfurization of high-sulfur oil from the Karazhanbas field of Kazakhstan with deep eutectic solvents.Heliyon. 2025 Jan 10;11(2):e41877. doi: 10.1016/j.heliyon.2025.e41877. eCollection 2025 Jan 30. Heliyon. 2025. PMID: 39906832 Free PMC article.
-
Comprehensive Molecular Dissection of Dermatophilus congolensis Genome and First Observation of tet(Z) Tetracycline Resistance.Int J Mol Sci. 2021 Jul 1;22(13):7128. doi: 10.3390/ijms22137128. Int J Mol Sci. 2021. PMID: 34281179 Free PMC article.
-
Microbacterium azadirachtae CNUC13 Enhances Salt Tolerance in Maize by Modulating Osmotic and Oxidative Stress.Biology (Basel). 2024 Apr 7;13(4):244. doi: 10.3390/biology13040244. Biology (Basel). 2024. PMID: 38666856 Free PMC article.
-
Characterization and Biosynthetic Regulation of Isoflavone Genistein in Deep-Sea Actinomycetes Microbacterium sp. B1075.Mar Drugs. 2024 Jun 13;22(6):276. doi: 10.3390/md22060276. Mar Drugs. 2024. PMID: 38921587 Free PMC article.
References
-
- Anderson M. J. (2001). A new method for non-parametric multivariate analysis of variance. Aust. Ecol. 26 32–46. 10.1111/j.1442-9993.2001.01070.pp.x - DOI
-
- Behrendt U., Ulrich A., Schumann P. (2001). Description of Microbacterium foliorum sp. nov. and Microbacterium phyllosphaerae sp. nov., isolated from the phyllosphere of grasses and the surface litter after mulching the sward, and reclassification of Aureobacterium resistens (Funke et al. 1998) as Microbacterium resistens comb. nov. Int. J. Syst. Evol. Microbiol. 51(Pt 4) 1267–1276. 10.1099/00207713-51-4-1267 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

