Prevalence and Distribution Characteristics of blaKPC-2 and blaNDM-1 Genes in Klebsiella pneumoniae
- PMID: 32903853
- PMCID: PMC7445519
- DOI: 10.2147/IDR.S253631
Prevalence and Distribution Characteristics of blaKPC-2 and blaNDM-1 Genes in Klebsiella pneumoniae
Abstract
Background: Carbapenem-resistant Klebsiella pneumoniae infections have caused major concern and posed a global threat to public health. As blaKPC-2 and blaNDM-1 genes are the most widely reported carbapenem resistant genes in K. pneumonia, it is crucial to study the prevalence and geographical distribution of these two genes for further understanding of their transmission mode and mechanism.
Purpose: Here, we investigated the prevalence and distribution of blaKPC-2 and blaNDM-1 genes in carbapenem-resistant K. pneumoniae strains from a tertiary hospital and from 1579 genomes available in the NCBI database, and further analyzed the possible core structure of blaKPC-2 or blaNDM-1 genes among global genome data.
Materials and methods: K. pneumoniae strains from a tertiary hospital in China during 2013-2018 were collected and their antimicrobial susceptibility testing for 28 antibiotics was determined. Whole-genome sequencing of carbapenem-resistant K. pneumoniae strains was used to investigate the genetic characterization. The phylogenetic relationships of these strains were investigated through pan-genome analysis. The epidemiology and distribution of blaKPC-2 and blaNDM-1 genes in K. pneumoniae based on 1579 global genomes and carbapenem-resistant K. pneumoniae strains from hospital were analyzed using bioinformatics. The possible core structure carrying blaKPC-2 or blaNDM-1 genes was investigated among global data.
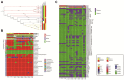
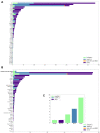
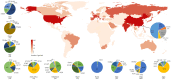
Results: A total of 19 carbapenem-resistant K. pneumoniae were isolated in a tertiary hospital. All isolates had a multi-resistant pattern and eight kinds of resistance genes. The phylogenetic analysis showed all isolates in the hospital were dominated by two lineages composed of ST11 and ST25, respectively. ST11 and ST25 were the major ST type carrying blaKPC-2 and blaNDM-1 genes, respectively. Among 1579 global genomes data, 147 known ST types (1195 genomes) have been identified, while ST258 (23.6%) and ST11 (22.1%) were the globally prevalent clones among the known ST types. Genetic environment analysis showed that the ISKpn7-dnaA/ISKpn27 -blaKPC-2-ISkpn6 and blaNDM-1-ble-trpf-nagA may be the core structure in the horizontal transfer of blaKPC-2 and blaNDM-1 , respectively. In addition, DNA transferase (hin) may be involved in the horizontal transfer or the expression of blaNDM-1 .
Conclusion: There was clonal transmission of carbapenem-resistant K. pneumoniae in the tertiary hospital in China. The prevalence and distribution of blaKPC-2 and blaNDM-1 varied by countries and were driven by different transposons carrying the core structure. This study shed light on the genetic environment of blaKPC-2 and blaNDM-1 and offered basic information about the mechanism of carbapenem-resistant K. pneumoniae dissemination.
Keywords: *blaNDM-1; Klebsiella pneumoniae; bioinformatics; blaKPC-2.
© 2020 Zhang et al.
Conflict of interest statement
All authors declare that they have no conflict of interest.
Figures

References
-
- Ripabelli G, Tamburro M, Guerrizio G, et al. Tracking multidrug-resistant Klebsiella pneumoniae from an Italian hospital: molecular epidemiology and surveillance by PFGE, RAPD and PCR-based resistance genes prevalence. Curr Microbiol. 2018;75(8):977–987. doi: 10.1007/s00284-018-1475-3 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

