The PhyloCode applied to Cintractiellales, a new order of smut fungi with unresolved phylogenetic relationships in the Ustilaginomycotina
- PMID: 32904025
- PMCID: PMC7451774
- DOI: 10.3114/fuse.2020.06.04
The PhyloCode applied to Cintractiellales, a new order of smut fungi with unresolved phylogenetic relationships in the Ustilaginomycotina
Abstract
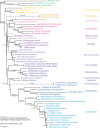
The PhyloCode is used to classify taxa based on their relation to a most recent common ancestor as recovered from a phylogenetic analysis. We examined the first specimen of Cintractiella (Ustilaginomycotina) collected from Australia and determined its systematic relationship to other Fungi. Three ribosomal DNA loci were analysed both with and without constraint to a phylogenomic hypothesis of the Ustilaginomycotina. Cintractiella did not share a most recent common ancestor with other orders of smut fungi. We used the PhyloCode to define the Cintractiellales, a monogeneric order with four species of Cintractiella, including C. scirpodendri sp. nov. on Scirpodendron ghaeri. The Cintractiellales may have shared a most recent common ancestor with the Malasseziomycetes, but are otherwise unresolved at the rank of class.
Keywords: Cyperaceae pathogens; ITS; LSU; fungal systematics; new taxa; obligate biotroph.
© 2020 Westerdijk Fungal Biodiversity Institute.
Figures


Similar articles
-
Broad Genomic Sampling Reveals a Smut Pathogenic Ancestry of the Fungal Clade Ustilaginomycotina.Mol Biol Evol. 2018 Aug 1;35(8):1840-1854. doi: 10.1093/molbev/msy072. Mol Biol Evol. 2018. PMID: 29771364
-
A new species of Cintractiella (Ustilaginales) from the volcanic island of Kosrae, Caroline Islands, Micronesia.MycoKeys. 2018 Nov 6;(42):1-6. doi: 10.3897/mycokeys.42.27231. eCollection 2018. MycoKeys. 2018. PMID: 30473621 Free PMC article.
-
On the Evolutionary History of Uleiella chilensis, a Smut Fungus Parasite of Araucaria araucana in South America: Uleiellales ord. nov. in Ustilaginomycetes.PLoS One. 2016 Jan 20;11(1):e0147107. doi: 10.1371/journal.pone.0147107. eCollection 2016. PLoS One. 2016. PMID: 26790149 Free PMC article.
-
A review of criticisms of phylogenetic nomenclature: is taxonomic freedom the fundamental issue?Biol Rev Camb Philos Soc. 2002 Feb;77(1):39-55. doi: 10.1017/s1464793101005802. Biol Rev Camb Philos Soc. 2002. PMID: 11911373 Review.
-
Smut fungi (Basidiomycota p.p., Ascomycota p.p.) of the world. novelties, selected examples, trends.Acta Microbiol Immunol Hung. 2008 Jun;55(2):91-109. doi: 10.1556/AMicr.55.2008.2.2. Acta Microbiol Immunol Hung. 2008. PMID: 18595315 Review.
Cited by
-
Fungal diversity notes 1512-1610: taxonomic and phylogenetic contributions on genera and species of fungal taxa.Fungal Divers. 2022;117(1):1-272. doi: 10.1007/s13225-022-00513-0. Epub 2023 Feb 23. Fungal Divers. 2022. PMID: 36852303 Free PMC article.
-
Impact of Plant Oil Supplementation on Lipid Production and Fatty Acid Composition in Cunninghamella elegans TISTR 3370.Microorganisms. 2024 May 15;12(5):992. doi: 10.3390/microorganisms12050992. Microorganisms. 2024. PMID: 38792821 Free PMC article.
References
-
- Aime MC, Matheny PB, Henk DA, et al. , (2006). An overview of the higher level classification of Pucciniomycotina based on combined analyses of nuclear large and small subunit rDNA sequences. Mycologia 98: 896–905. - PubMed
-
- Albu S, Toome M, Aime MC. (2015). Violaceomyces palustris gen. et sp. nov. and a new monotypic lineage, Violaceomycetales ord. nov. in Ustilaginomycetes. Mycologia 107: 1193–1204. - PubMed
-
- Bauer R, Oberwinkler F, Vánky K. (1997). Ultrastructural markers and systematics in smut fungi and allied taxa. Canadian Journal of Botany 75: 1273–1314.
LinkOut - more resources
Full Text Sources
