Response of Soybean Root to Phosphorus Deficiency under Sucrose Feeding: Insight from Morphological and Metabolome Characterizations
- PMID: 32904516
- PMCID: PMC7456465
- DOI: 10.1155/2020/2148032
Response of Soybean Root to Phosphorus Deficiency under Sucrose Feeding: Insight from Morphological and Metabolome Characterizations
Abstract
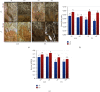
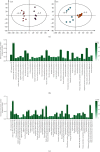
Phosphorus (P) is one the least available essential plant macronutrients in soils that is a major constraint on plant growth. Soybean (Glycine max L.) production is often limited due to low P availability. The better management of P deficiency requires improvement of soybean's P use efficiency. Sugars are implicated in P starvation responses, and a complete understanding of the role of sucrose together with P in coordinating P starvation responses is missing in soybean. This study explored global metabolomic changes in previously screened low-P-tolerant (Liaodou, L13) and low-P-sensitive (Tiefeng 3, T3) soybean genotypes by liquid chromatography coupled mass spectrometry. We also studied the root morphological response to sucrose application (1%) in P-starved soybean genotypes against normal P supply. Root morphology in L13 genotype has significantly improved P starvation responses as compared to the T3 genotype. Exogenous sucrose application greatly affected root length, root volume, and root surface area in L13 genotype while low-P-sensitive genotype, i.e., T3, only responded by increasing number of lateral roots. Root : shoot ratio increased after sucrose treatment regardless of P conditions, in both genotypes. T3 showed a relatively higher number of differentially accumulated metabolites between P-starved and normal P conditions as compared to L13 genotype. Common metabolites accumulated under the influence of sucrose were 5-O-methylembelin, D-glucuronic acid, and N-acetyl-L-phenylalanine. We have discussed the possible roles of the pathways associated with these metabolites. The differentially accumulated metabolites between both genotypes under the influence of sucrose are also discussed. These results are important to further explore the role of sucrose in the observed pathways. Especially, our results are relevant to formulate strategies for improving P efficiency of soybean genotypes with different P efficiencies.
Copyright © 2020 Ahui Yang et al.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Integration of metabolome and transcriptome analyses highlights soybean roots responding to phosphorus deficiency by modulating phosphorylated metabolite processes.Plant Physiol Biochem. 2019 Jun;139:697-706. doi: 10.1016/j.plaphy.2019.04.033. Epub 2019 Apr 26. Plant Physiol Biochem. 2019. PMID: 31054472
-
Up-regulating GmETO1 improves phosphorus uptake and use efficiency by promoting root growth in soybean.Plant Cell Environ. 2020 Sep;43(9):2080-2094. doi: 10.1111/pce.13816. Epub 2020 Jul 16. Plant Cell Environ. 2020. PMID: 32515009
-
Microarray analysis of iron deficiency chlorosis in near-isogenic soybean lines.BMC Genomics. 2007 Dec 21;8:476. doi: 10.1186/1471-2164-8-476. BMC Genomics. 2007. PMID: 18154662 Free PMC article.
-
Mechanisms Underlying Soybean Response to Phosphorus Deficiency through Integration of Omics Analysis.Int J Mol Sci. 2022 Apr 21;23(9):4592. doi: 10.3390/ijms23094592. Int J Mol Sci. 2022. PMID: 35562981 Free PMC article. Review.
-
Sucrose transport in the phloem: integrating root responses to phosphorus starvation.J Exp Bot. 2008;59(1):93-109. doi: 10.1093/jxb/erm221. J Exp Bot. 2008. PMID: 18212031 Review.
Cited by
-
Metabolomics as a Prospective Tool for Soybean (Glycine max) Crop Improvement.Curr Issues Mol Biol. 2022 Sep 12;44(9):4181-4196. doi: 10.3390/cimb44090287. Curr Issues Mol Biol. 2022. PMID: 36135199 Free PMC article. Review.
-
Proteome Analysis of the Soybean Nodule Phosphorus Response Mechanism and Characterization of Stress-Induced Ribosome Structural and Protein Expression Changes.Front Plant Sci. 2022 Jun 9;13:908889. doi: 10.3389/fpls.2022.908889. eCollection 2022. Front Plant Sci. 2022. PMID: 35755677 Free PMC article.
-
Functional analysis of ZmG6PE reveals its role in responses to low-phosphorus stress and regulation of grain yield in maize.Front Plant Sci. 2023 Nov 9;14:1286699. doi: 10.3389/fpls.2023.1286699. eCollection 2023. Front Plant Sci. 2023. PMID: 38023907 Free PMC article.
-
Proteome characterization of two contrasting soybean genotypes in response to different phosphorus treatments.AoB Plants. 2021 Apr 14;13(3):plab019. doi: 10.1093/aobpla/plab019. eCollection 2021 Jun. AoB Plants. 2021. PMID: 34150189 Free PMC article.
-
Screening and identification of evaluation indicators of low phosphorus tolerant germplasm in Gleditsia sinensis Lam.Sci Rep. 2024 Dec 30;14(1):31716. doi: 10.1038/s41598-024-82071-w. Sci Rep. 2024. PMID: 39738241 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

