Analysis of sequence diversity and selection pressure in HIV-1 clade C gp41 from India
- PMID: 32904888
- PMCID: PMC7458999
- DOI: 10.1007/s13337-020-00595-x
Analysis of sequence diversity and selection pressure in HIV-1 clade C gp41 from India
Abstract
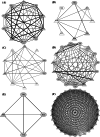
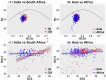
Evaluation of viral diversity is critical for the rational design of treatment modalities against Human immunodeficiency virus (HIV). Predominated by HIV-1 clade C (HIV-1C), the epidemic in India represents the third largest population infected with HIV-1 globally. Glycoprotein 41 (gp41) is critical for viral replication and is a target for the design of therapeutic strategies. However, documentation of viral diversity of gp41 gene in infected individuals from India remains limited. Present study employed high throughput sequencing to examine variation in gp41 amplicons generated from blood derived viruses in 24 HIV-1C infected individuals from Mumbai, India. Sequence diversity profiles were documented in different functional domains of gp41. Furthermore, through a meta-analysis approach, all reported gp41 sequences from India (N = 70) were compared with those from South Africa (N = 126), country with the largest HIV epidemic globally, also predominated by HIV-1C. A total of 44 positions displayed statistically significant differential (p < 0.05) Shannon entropy in the two regions. This comparison also identified 11 codon sites undergoing distinct selection, 8 of which remained differentially selected in an extended comparison of data from Asia (N = 137) and Africa(N = 383). Assessment of correlated mutation networks associated with differentially selected residues revealed common as well as distinct interaction networks. Furthermore, codon usage analysis revealed 17 differentially selected codons (Mann-Whitney test, p < 0.001) in Asia and Africa. Dissimilar trends in GC content across codon positions were also observed. In depth understanding of these divergent evolutionary signatures through extended analysis with larger data-sets would assist development of effective interventions being considered for HIV-1C.
Keywords: Codon usage; Evolution; HIV-1C; India; Viral variation; gp41.
© Indian Virological Society 2020.
Figures



References
-
- Bandawe GP, Martin DP, Treurnicht F, Mlisana K, Karim SSA, Williamson C, et al. Conserved positive selection signals in gp41 across multiple subtypes and difference in selection signals detectable in gp41 sequences sampled during acute and chronic HIV-1 subtype C infection. Virol J. 2008;5:141. doi: 10.1186/1743-422X-5-141. - DOI - PMC - PubMed
-
- Bellamy-McIntyre AK, Lay C-S, Baär S, Maerz AL, Talbo GH, Drummer HE, et al. Functional links between the fusion peptide-proximal polar segment and membrane-proximal region of human immunodeficiency virus gp41 in distinct phases of membrane fusion. J Biol Chem. 2007;282:23104–23116. doi: 10.1074/jbc.M703485200. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

